Abstract
Background
Oxidative stress is now one of the accepted theories of vitiligo development. Nuclear factor erythroid-2-related factor 2 (Nrf2) regulates the expression of antioxidant proteins.
Objective
This work aimed to evaluate the association of Nrf2 gene polymorphisms with the susceptibility to vitiligo among a sample of Egyptian patients with vitiligo.
Methods
This case-control study included 100 patients with vitiligo and 50 healthy matched volunteers serving as a control group. Genotyping was carried out by real-time polymerase chain reaction.
Results
The frequencies of TT, CT, and combined (TT+CT) genotypes and the T allele of Nrf2 (rs35652124) were significantly increased in the studied patients with vitiligo relative to the healthy controls (p<0.001, p=0.012, p<0.001 and p<0.001, respectively). There was a nonsignificant difference between patients and controls regarding Nrf2 (rs6721961) genotypes. However, the T allele of Nrf2 (rs6721961) was significantly predominant in the studied patients compared to in the controls (p=0.029). Among the studied criteria, the T allele of Nrf2 (rs6721961) was predominant in patients with a marginal type of repigmentation (p=0.022), while the G allele of the same single-nucleotide polymorphism was associated with a higher body mass index value (p=0.034). One hundred percent of patients with vitiligo with the Nrf2 (rs6721961) GT genotype had a progressive disease course (p=0.015).
Conclusion
Nrf2 (−617 T/G) and (−653 T/C) polymorphism might play a role in patient susceptibility to vitiligo and modify the clinical presentation of the disease.
Keywords: Gene polymorphism, nuclear factor erythroid-2-related factor 2, vitiligo
Vitiligo is the most common acquired depigmenting disorder affecting genetically predisposed individuals.1 How vitiligo develops is not yet well established; however, autoimmune, oxidative stress, neural, and biochemical-based theories have been put forth.2 Reactive oxygen species (ROS) created via different pathways might contribute to the destruction of melanocytes either via direct damage or through increased melanocyte-specific autoantibody production.3
Nuclear factor erythroid-2-related factor 2 (Nrf2) is a basic leucine zipper protein that plays a role in regulating the expression of antioxidant proteins that protect cells against the damaging effects of oxidative stress.4,5
Nrf2 gene polymorphisms, especially those affecting the promoter region, can impair Nrf2 protein expression and activity. Two polymorphisms in the promoter region—a T to C substitution at position −653 (Nrf2 rs35652124) and a G to T substitution at −617 (Nrf2 rs6721961)—are described and known to be associated with different pathologic conditions.6
This study aimed to investigate the association between Nrf2 (rs35652124) and (rs6721961) gene polymorphisms and the susceptibility to vitiligo and to assess the effects of different genotypes on the clinical aspects of the disease.
METHODS
The study included 100 patients suffering from different clinical presentations of vitiligo and 50 healthy, vitiligo-free individuals from the Dermatology Outpatient Clinic of Benha University Hospital. The study was approved by the ethical committee of human research of the Faculty of Medicine at Benha University. After discussing the study's risks, benefits, and other aspects with all participants, written informed consent was obtained from each participant before sample collection. The history of the present illness, related medical conditions, and associated diseases were discussed in details with all patients. Patients with other inflammatory or autoimmune cutaneous or systemic disorders were excluded from this study.
DNA extraction and genotyping. Genomic DNA was extracted from peripheral blood samples from all participants. Genomic DNA was isolated using the GeneJET genomic DNA purification kit (cat. no. K0721; Qiagen, Hilden, Germany). The purity and concentration of DNA were evaluated spectrophotometrically using Nanodrop (Thermo Fisher Scientific, Waltham, Massachusetts). The quality of DNA was assessed with the A260/280 ratio. Genotyping was performed using the Taqman single nucleotide polymorphism (SNP) ready-made assay (Qiagen), which includes the TaqMan Universal Master Mix II (cat. no. 4440043) and TaqMan SNP human genotyping assay (cat. no. 4351379). The TaqMan genotyping assay used forward primer, reverse primer, and two probes as follows: rs35652124, forward 5-CCT TGC CCTGCT TTT ATC TC-3 and reverse 5-CTT CTC CGT TTG CCT TTG AC-3; rs6721961, forward 5-GAA AGG CGT TGG TGT AGG AG-3 and reverse 5-GAA TGGAGA CAC GTG GGA GT-3; normal probe, (5'-3') VIC-ACTCCTTTCACCTATTCCCAAGGCCT-MGB-NFQ; and mutant probe, (5'-3') FAM-CAGCTACACCTGTATGTAGGCTAGA MGB-NFQ. The probes were designed with a minor groove binder and nonfluorescent quencher at the 3' end, whereas the 5' end contained the fluorescence reporter dyes 2'-chloro-7'-phenyl-1,4-dichloro-6-carboxyfluorescein (VIC) or 6-carboxyfluorescein (FAM). The wild-type probe was labeled with VIC dye, while the variant probe was labeled with FAM dye. Polymerase chain reaction was performed in a volume of 20μL using the Rotorgene real-time polymerase chain reaction system (Qiagens). Thermal cycling conditions were as follows: 60°C for 30 seconds, 95°C for 10 minutes, 40 cycles of denaturation, 95°C for 15 seconds, and 60°C for one minute.
Statistical analysis. Data were coded and entered using the Statistical Package for the Social Sciences version 25 software program (IBM Corporation, Armonk, New York). Data were summarized using mean, standard deviation, median, minimum, and maximum values if quantitative data and frequency (count) and relative frequency (percentage) of categorical data. Comparisons between quantitative variables were done using the nonparametric Kruskal–Wallis and Mann–Whitney U tests.
For comparing categorical data, the chi-squared test was performed. Fisher’s exact test was used instead when the expected frequency was less than 5. Genotype and allele frequencies were compared between the disease and the control groups. Odds ratios with 95% confidence intervals were calculated using binary logistic regression, and p-values of less than 0.05 were considered to be statistically significant.
RESULTS
The patient and control groups were matched regarding age (23.79±15.87 vs. 24.46±9.12 years; p=0.147), sex (male:female ratios of 68:32 among patients and 30:20 among control participants; p=0.332), and body mass index (BMI) (24.41±5.57 kg/m2 vs. 25.49±4.40 kg/m2; p=0.094).
The mean disease duration was 6.68±6.39 years. A stressful condition was associated with disease onset in 51 percent of cases. Koebnerization was positive in 36 percent of the patients with vitiligo. Hearing and vision impairments were found in 10 percent and eight percent of the patients, respectively. Regarding the clinical type of vitiligo in the studied sample, 86 patients had nonsegmental vitiligo and 14 patients presented with segmental vitiligo. Leukotrichia was observed in 37 cases. The mean Vitiligo Area Scoring Index score was 13.16±24.16.
Applying the Hardy–Weinberg equation, it was revealed that Nrf2 genotypes and alleles frequency in the patient and control groups in both studied SNPs were in Hardy Weinberg equilibrium. The TT and CT genotypes and T allele of the Nrf2 gene (rs35652124) and T allele of Nrf2 (rs6721961) increase the risk of vitiligo significantly (Table 1).
TABLE 1.
Genotypes and alleles of the studied SNPs
| GENOTYPES AND ALLELES | CASES (N=100) | CONTROL (N=50) | P-VALUE | ODDS RATIO | 95% CONFIDENCE INTERVAL | |
|---|---|---|---|---|---|---|
| N (%) | N (%) | UPPER | LOWER | |||
| Nrf2 (rs35652124) | ||||||
| TT (mutant) | 48 (48.0) | 10 (20.0) | <0.001 | 7.040 | 2.734 | 18.130 |
| CT (heterozygous) | 37 (37.0) | 18 (36.0) | 0.012 | 3.015 | 1.27 | 7.159 |
| CC (wild) | 15 (15.0) | 22 (44.0) | Reference | |||
| TT (mutant) + CT (heterozygous) | 85 (85.0) | 28 (56.0) | <0.001 | 4.452 | 2.035 | 9.742 |
| allele T | 133 (66.5) | 38 (38.0) | <0.001 | 3.239 | 1.966 | 5.336 |
| allele C | 67 (33.5) | 62 (62.0) | Reference | |||
| Nrf2 (rs6721961) | ||||||
| TT (mutant) | 20 (20.0) | 5 (10.0) | 0.062 | 2.842 | 0.949 | 8.514 |
| GT (heterozygous) | 42 (42.0) | 18 (36.0) | 0.181 | 1.658 | 0.791 | 3.476 |
| GG (wild) | 38 (38.0) | 27 (54.0) | Reference | |||
| TT+GT | 62 (62.0) | 23 (46.0) | 0.064 | 1.915 | 0.963 | 3.808 |
| allele T | 82 (41.0) | 28 (28.0) | 0.029 | 1.787 | 1.063 | 3.004 |
| allele G | 118 (59.0) | 72 (72.0) | Reference | |||
| Haplotype | ||||||
| rs35652124 allele T + rs6721961 allele T | 71 (35.5) | 18 (18.0) | <0.001 | 3.663 | 1.931 | 6.948 |
| rs35652124 allele T + rs6721961 allele G | 62 (31.0) | 20 (20.0) | 0.001 | 2.879 | 1.534 | 5.403 |
| rs35652124 allele C + rs6721961 allele T | 11 (5.5) | 10 (10.0) | 0.965 | 1.021 | 0.401 | 2.604 |
| rs35652124 allele C + rs6721961 allele G | 56 (28.0) | 52 (52.0) | Reference | |||
Among the studied variables, there was a nonsignificant relationship between the genotypes and alleles of the studied SNPs, except for the significant association between the T allele of rs6721961 and the marginal type of repigmentation (Figure 1) and between the G allele of the same SNP and the BMI (Figure 2).
FIGURE 1.
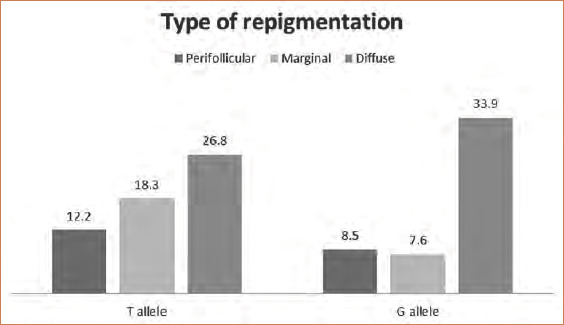
Relationship between different Nrf2 (rs6721961) alleles and types of repigmentation
FIGURE 2.
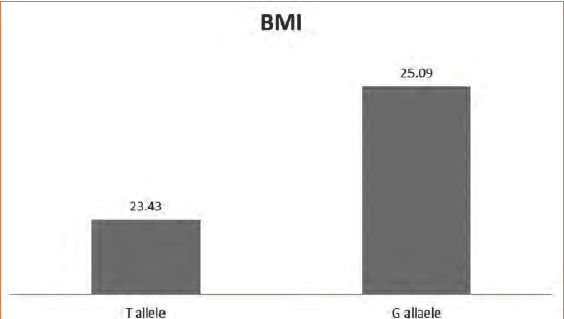
Relationship between Nrf2 (rs6721961) alleles and body mass index
DISCUSSION
Nrf2 and its negative regulator protein, Keap1, play a crucial antioxidant role, protecting all body cells against the harmful effects exerted by the oxidative stress.7 Moreover, activating Nrf2 might be a promising therapeutic option in managing many inflammatory conditions in which oxidative stress might be involved in the pathogenesis.8
Many SNPs in Keap 1 and Nrf2 have been investigated and their association with many inflammatory diseases are now documented.9 Of particular importance, the two promotor regions of the gene SNP are the rs35652124 and the rs6721961 polymorphisms. The ancestral alleles at Nrf2 (rs35652124) and (rs6721961) are the C allele and the G allele, respectively. Both ancestral alleles are associated with greater Nrf2 activity.6,10
In the present work, a significant link between Nrf2 (rs35652124) polymorphism and vitiligo was observed, while there was a nonsignificant difference between patients and controls regarding Nrf2 (rs6721961) genotypes. However, the T allele of Nrf2 (rs6721961) increased the risk of vitiligo.
Song et al6 investigated the association of the same gene SNPs with vitiligo in a Chinese population (1,136 patients and 1,200 controls). In accordance with the present work, these authors reported an association between the Nrf2 (rs35652124) TT and CT genotypes and an increased risk of vitiligo. However, they didn not observe a significant difference between patients and controls regarding Nrf2 (rs6721961) genotypes or alleles. The discrepancy between the results of these two studies might be explained by the difference in sample size, ethnic variations, and different genetic and racial backgrounds.
In the present study, the T allele of Nrf2 (rs6721961) was significantly more predominant in vitiligo patients with the marginal type of repigmentation. Selvan et al11 reported that the marginal pattern was predominant in both narrowband ultraviolet B and excimer laser-treated patients. Moreover, Yang et al12 reported that patients with vitiligo treated with topical therapies mostly developed marginal repigmentation. However, Gan et al13 found that the most common pattern of repigmentation was the perifollicular pattern, followed by the diffuse, combined, and marginal repigmentation types, respectively. During six months of follow-up after cessation of the applied therapeutic lines, Gan et al13 also detected that marginal repigmentation was the most stable form. From these findings, we can predict that carriers of the T allele of Nrf2 (6721961) would have a good prognosis.
The G allele of Nrf2 (rs6721961), known to be associated with high Nrf2 activity, was associated with higher BMI in the studied patients. This finding was interesting because Shin et al14 demonstrated the effects of Nrf2 in preventing fat accumulation and body weight gain. However, this finding could be explained by the exhaustion of Nrf2 function due to its constant activation.15
The incidence of gastrointestinal diseases (e.g., ulcerative colitis, celiac disease) in vitiligo patients is higher than that in the normal population.16 Arisawa et al17 reported that the Nrf2 (rs35652124) C allele is protective against gastrointestinal diseases, and patients from a Japanese population heterozygous at locus Nrf2 (rs35652124) CT were more susceptible to ulcerative colitis. This could provide a genetic explanation for the association between these conditions.
CONCLUSION
Nrf2 (−617 T/G) and (−653 T/C) polymorphism might place a role in patients’ susceptibility to vitiligo and modify the clinical presentation of the disease. Based on our results, we recommend further studies to investigate the Nrf2 expression in vitiligo lesions as well as the relationship between different Nrf2 (−617 T/G) and (−653 T/C) genotypes with the expression levels.
REFERENCES
- Saleem MD, Oussedik E, Schoch JJ et al. Acquired disorders with depigmentation: a systematic approach to vitiliginoid conditions. J Am Acad Dermatol. 2019;80(5):1215–1231.e6. doi: 10.1016/j.jaad.2018.03.063. [DOI] [PubMed] [Google Scholar]
- Gianfaldoni S, Tchernev G, Wollina U et al. Vitiligo in children: a better understanding of the disease. Maced J Med Sci. 2018;6(1):181–184. doi: 10.3889/oamjms.2018.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li S, Li C. Perspectives of new advances in the pathogenesis of vitiligo: from oxidative stress to autoimmunity. Med Sci Monit. 2019;25:1017–1023. doi: 10.12659/MSM.914898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu X, Long M et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. 2016;8:334ra51. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Yamamoto M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J Biol Chem. 2017;292(41):16817–16824. doi: 10.1074/jbc.R117.800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Li K, Liu L et al. Genetic polymorphism of the Nrf2 promoter region is associated with vitiligo risk in Han Chinese populations. J Cell Mol Med. 2016;20(10):1840–1850. doi: 10.1111/jcmm.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama Y, Mitsuda Y, Hamajima N, Niwa T. Polymorphisms of Nrf2, an antioxidative gene, are associated with blood pressure in Japanese. Nagoya J Med Sci. 2014;76(1–2):113–120. [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Niu T, Liu H, Tay FR. Protection against HEMA-induced mitochondrial injury in vitro by Nrf2 activation. Oxid Med Cell Longev. 2019;2019:3501059. doi: 10.1155/2019/3501059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamodharan U, Ponjayanthi B, Sireesh D et al. Association of single-nucleotide polymorphisms of the KEAP1 gene with the risk of various human diseases and its functional impact using in silico analysis. Pharmacol Res. 2018;137:205–218. doi: 10.1016/j.phrs.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Marzec JM, Christie JD, Reddy SP et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007;21(9):2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- Selvan BS, Thadeus J, Anandan H. Pattern of repigmentation in the treatment of vitiligo vulgaris with NBUVB therapy. Int J Contemp Med Res. 2018;5(5):E1–E3. [Google Scholar]
- Yang K, Xiong X, Pallavi G et al. The early repigmentation pattern of vitiligo is related to the source of melanocytes and by the choice of therapy: a retrospective cohort study. Int J Dermatol. 2018;57(3):324–331. doi: 10.1111/ijd.13878. [DOI] [PubMed] [Google Scholar]
- Gan EY, Eleftheriadou V, Esmat S et al. Repigmentation in vitiligo: position paper of the Vitiligo Global Issues Consensus Conference. Pigment Cell Melanoma Res. 2017;30(1):28–40. doi: 10.1111/pcmr.12561. [DOI] [PubMed] [Google Scholar]
- Shin S, Wakabayashi J, Yates MS et al. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. 2009;620(1–3):138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HA, Lee IK. The role of Nrf 2: adipocyte differentiation, obesity, and insulin resistance. Oxid Med Cell Longev. 2013;2013:184598. doi: 10.1155/2013/184598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashankar D, Prendiville J, Israel DM. Vitiligo and Crohn's disease in children. J Pediatr Gastroenterol Nutr. 1999;28(2):227–229. doi: 10.1097/00005176-199902000-00029. [DOI] [PubMed] [Google Scholar]
- Arisawa T, Tahara T, Shibata T et al. The influence of promoter polymorphism of nuclear factor-erythroid 2-related factor 2 gene on the aberrant DNA methylation in gastric epithelium. Oncol Rep. 2008;19(1):211–216. [PubMed] [Google Scholar]


