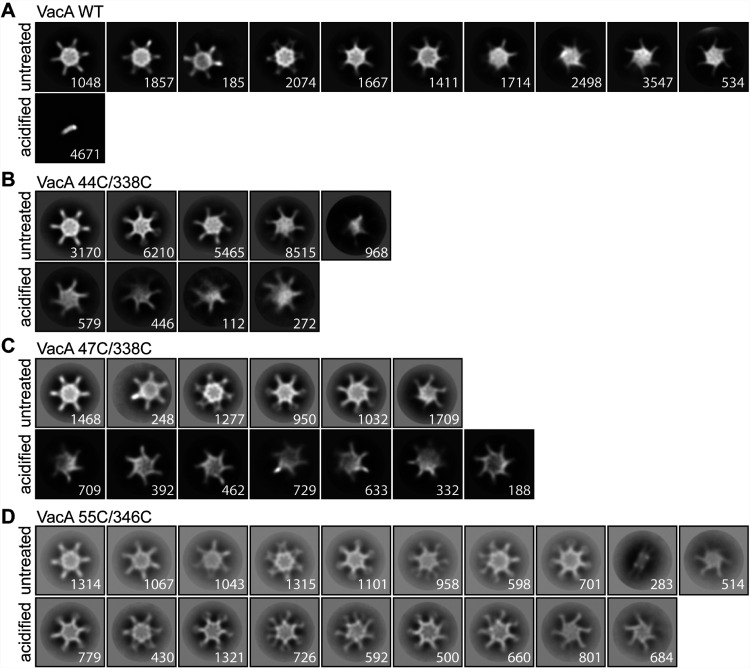
FIG 6.
2D class averages of VacA proteins visualized by negative-stain EM. WT VacA and the indicated VacA mutant proteins containing paired cysteine substitutions were left untreated or were acidified and then were analyzed by negative-stain EM. 2D class averages were generated as described in Materials and Methods. Representative class averages of each protein are shown. WT VacA disassembled into monomers at low pH, whereas the mutants retained an oligomeric structure under these conditions. Numbers of particles are shown in the bottom right corner of each class. Box size, 468 Å.