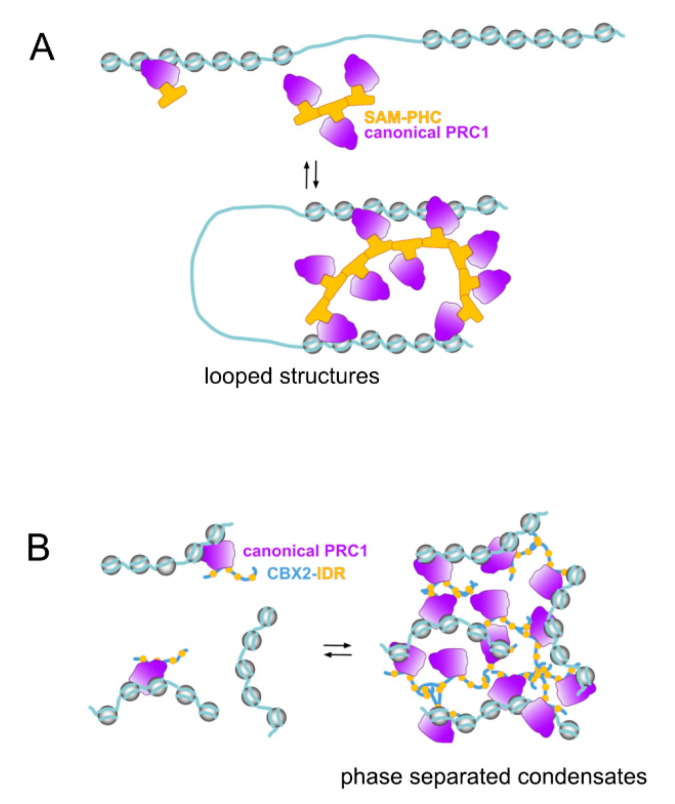
Figure 2.
Polycomb and genomic compartmentalization functions. Characteristic of Polycomb activities is the ability to organize subsets of repressed loci in genomic structures where loci are clustered through singular protein–protein interactions between select canonical PRC1 subunits. (A) architectural functions mediated by SAM domain-containing subunits (Polyhomeotic paralogs, for example) underlie the formation of looped structures through contacts between genomic sites separated. The ability of every SAM domain to make contacts with two SAM domains is key to the stability of these structures. (B) an alternative source of clustered structures implies internally disorganized regions (IDRs) as those in CBX2/M33 in mammals, or PSC in Drosophila. Promiscuous contacts among IDRs, after achieving very high local concentrations gives rises to phase-separated condensates where the “trapped” genomic sites are physically separated from other nuclear components. Although transcriptional repression correlates with Polycomb condensates, these structures are not gene inactivation-specific (see [127]).