Fig. 2 |. Biochemical and biophysical characterization of DNMT1-selective inhibitors.
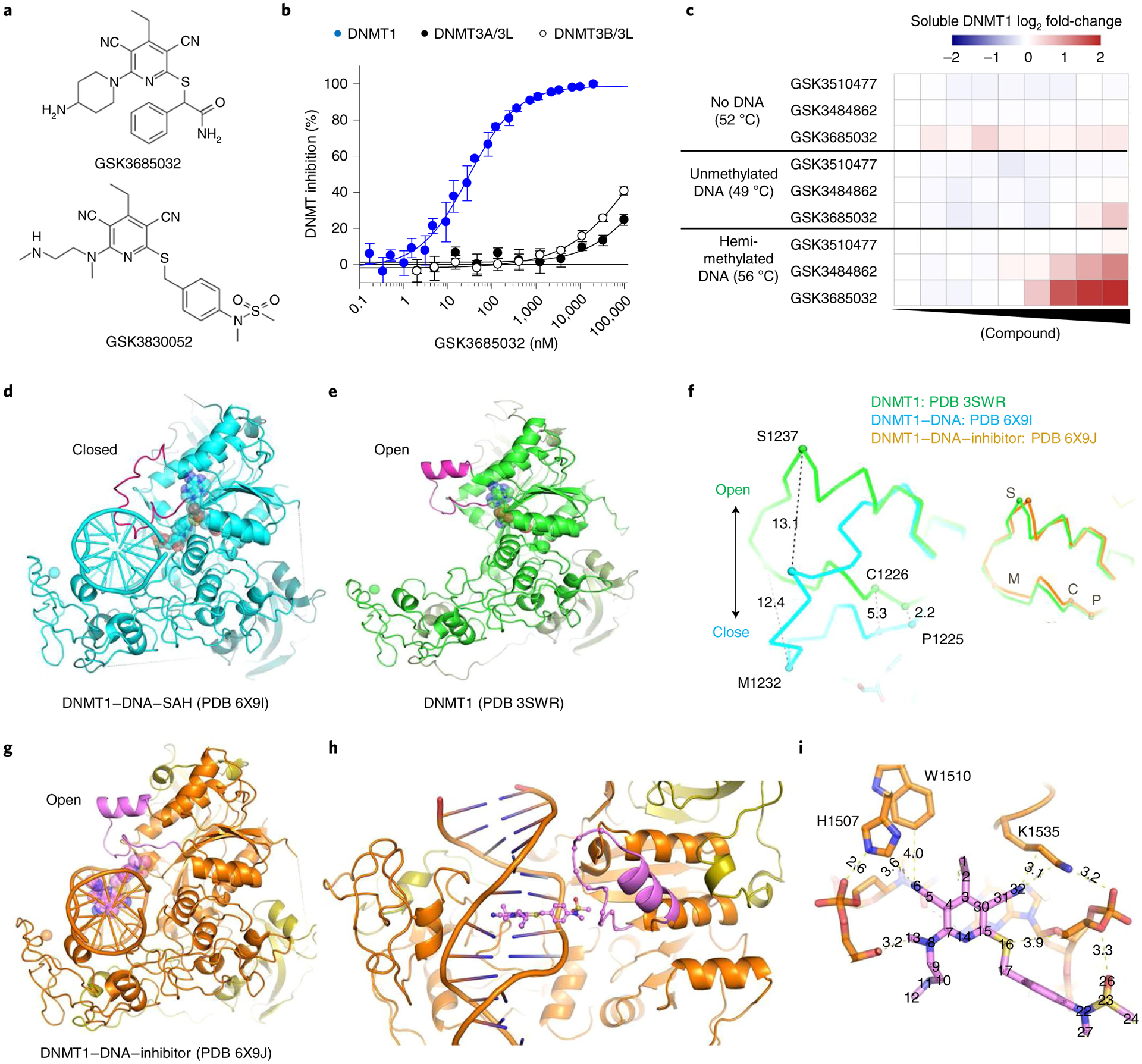
a, GSK3685032 and GSK3830052 structures. b, GSK3685032 dose–response curves for the DNMT family generated using a radioactive SPA (n = 4 biologically independent samples; average ± s.d.). c, Thermal stabilization of recombinant DNMT1 (601–1600) upon compound treatment (0.13–50,000 nM) in the presence of 40-mer hemi-methylated DNA or unmethylated DNA, or in the absence of DNA. d, Structure of DNMT1–DNA where the DNA contains zebularine in place of the target cytidine (PDB 6X9I). e, DNMT1 in the absence of bound DNA (PDB 3SWR). f, Conformational changes of the active-site loop (residues 1224–1245). Left panel, DNMT1 with and without DNA. Right panel, DNMT1 in the absence of DNA or as part of the DNMT1–DNA–inhibitor complex. C, Cys1226; M, Met1232; P, Pro1225; S, Ser1237. g, Structure of DNMT1–DNA complex in the presence of GSK3830052 (PDB 6X9J). h, Close-up view of the inhibitor in the minor groove. i, Inhibitor-mediated interactions with residues in the DNA major groove side.
