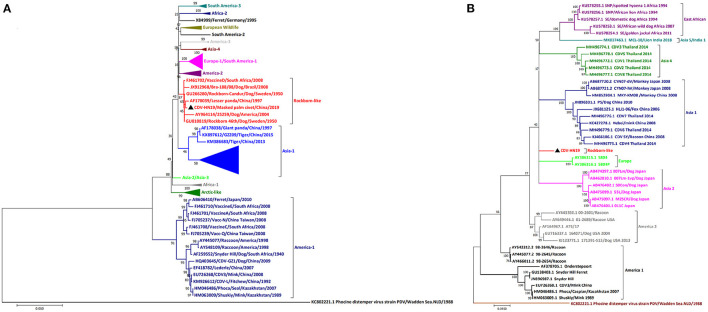
Figure 2.
Phylogenetic analyses of CDV HN19 strain. (A) Phylogenetic analyses of the nucleotide sequences of the complete genome of CDV HN19 strain. (B) Phylogenetic analyses of H gene nucleotide sequences of CDV HN19. Evolutionary history was inferred using the maximum likelihood method with the Tamura-Nei model and gamma-distributed rate heterogeneity in MEGA 7. The percentage of replicates in which the associated virus clustered together in the bootstrap test (1,000 replicates) is shown next to the branch in each tree. The strain isolated in this study is identified by ▲. The percentage bootstrap support is indicated by the value at each node. Scale bar denotes nucleotide substitutions per site.