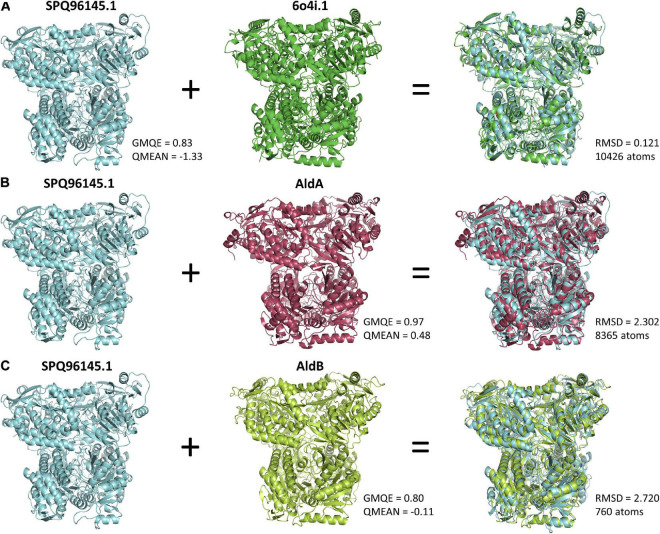
FIGURE 5.
Predicted 3D structure of aldehyde dehydrogenases. (A) The aldehyde dehydrogenase from P. brassicae (SPQ96145.1) was predicted using Alpha-aminoadipic semialdehyde dehydrogenase (STML ID:6o4i.1.A – second structure from left to right) using Swiss-model. The structural alignment of the two structures can be seen on the third column. (B,C) Models for aldehyde dehydrogenases A and B (AldA and AldB -second column) from P. syringae which are involved in IAA synthesis, were aligned with the P. brassicae model, to produce the structural alignments seen on column 3. RMSD, root mean square deviation of all aligned atoms between the two structures, where a value of 0 is a perfect alignment. The number of aligned atoms is also reported for the three structural alignments. A Global Model Quality Estimation (GMQE) score closer to 1 represents more accuracy in the models, and a QMEAN value above –4 is indicative of good quality of the structure (https://swissmodel.expasy.org/docs/help).