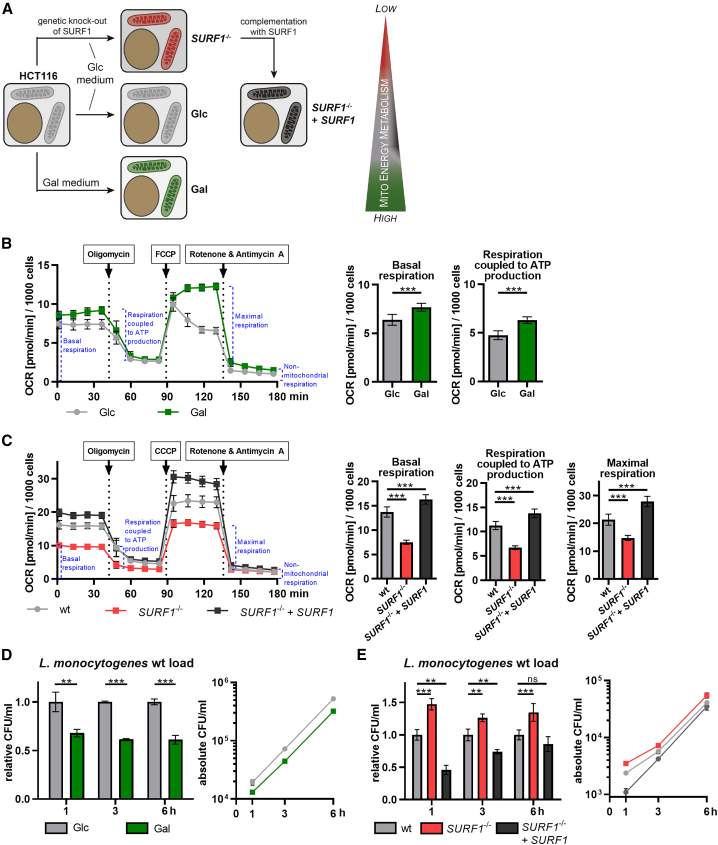
Figure 1.
Mitochondrial energy metabolism affects L. monocytogenes infection
(A) Overview of the approaches used to generate cellular models with increased or decreased mitochondrial energy metabolism. HCT116 cells grown in galactose (Gal)-containing medium are energetically dependent on mitochondrial respiration (green mitochondria), whereas cells grown in glucose (Glc)-containing medium are mainly glycolytic and rely less on mitochondrial respiration (light gray mitochondria). Cells depleted for the mitochondrial protein SURF1 (SURF1−/− cells) show decreased mitochondrial respiration (red mitochondria), which can be rescued by reintroducing the SURF1 gene (SURF1−/− + SURF1 cells, dark gray mitochondria). The wedge indicates the level of dependency on mitochondrial respiration for energy production for each cell model.
(B and C) Oxygen consumption rate (OCR; in picomoles per minute) of HCT116 Glc and Gal cells (B) and of HCT116 WT, SURF1−/−, and SURF1−/− + SURF1 cells (C) monitored in a Seahorse XFe96 analyzer. Three independent experiments were performed and data from one representative experiment with six biological replicates per condition are shown as mean ± standard deviation (SD) for each time point. The rates of basal respiration, respiration coupled to ATP production, and maximal respiration were statistically evaluated by two-tailed t tests (B) and one-way ANOVA with Dunnett’s post hoc test (C) (∗∗∗p < 0.001).
(D and E) Intracellular bacterial load in HCT116 Glc and Gal cells (D) and in HCT116 WT, SURF1−/−, and SURF1−/− + SURF1 infected with WT L. monocytogenes EGDe (MOI, 20). The left panel shows values for Gal cells relative to Glc cells and values for SURF1−/− and SURF1−/− + SURF1 cells relative to WT cells, and the right panel shows the absolute quantification (CFU/mL), for each time point. Three independent experiments were performed, and for both panels, one representative experiment with three biological replicates is shown as mean ± SD. Statistical significances were calculated by multiple t tests (D) and one-way ANOVA with multiple comparisons (E); both were further evaluated by the false discovery rate approach of Benjamini, Krieger ,and Yekutieli, with Q = 1% (ns, not significant; ∗∗p < 0.01; ∗∗∗p < 0.001).