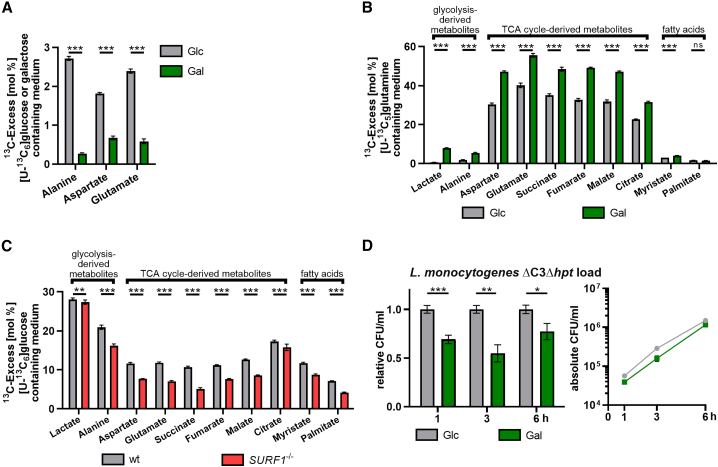
Figure 2.
Characterization of changes in mitochondrial respiration induced by medium formulation or genetic ablation of the mitochondrial protein SURF1
(A) 13C-Excess in proteinogenic amino acids from HCT116 Glc and Gal cells labeled with [U-13C6]Glc or [U-13C6]Gal, respectively, as quantified by gas chromatography-mass spectrometry (GC-MS). Shown are the mean ± SD (cumulative biological and technical errors) of one experiment with two biological replicates, where each one is analyzed three times in technical replicates. The differences between HCT116 Glc and Gal cells were statistically evaluated by two-tailed t tests (∗∗∗p < 0.001).
(B) 13C-Excess in cytosolic metabolites from HCT116 Glc and Gal cells labeled with [U-13C5]glutamine as quantified by GC-MS. Experimental setup and statistical analysis were performed as in (A) (∗∗∗p < 0.001).
(C) 13C-Excess in cytosolic metabolites from HCT116 WT and SURF1−/− cells labeled with [U-13C6]Glc as quantified by GC-MS. Experimental setup and statistical analysis were performed as in (A) (∗∗p < 0.01; ∗∗∗p < 0.001).
(D) Intracellular bacterial load in HCT116 Glc and Gal cells infected with L. monocytogenes EGDe ΔC3Δhpt (MOI, 20). The left panel shows values for Gal cells relative to Glc cells, and the right panel shows the absolute quantification (CFU/ml) for each time point. Three independent experiments were performed, and for both panels, data from one representative experiment with three biological replicates are shown as mean ± SD. Statistical significances were determined by multiple t tests and evaluated by the false discovery rate approach of Benjamini, Krieger, and Yekutieli, with Q = 1% (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).