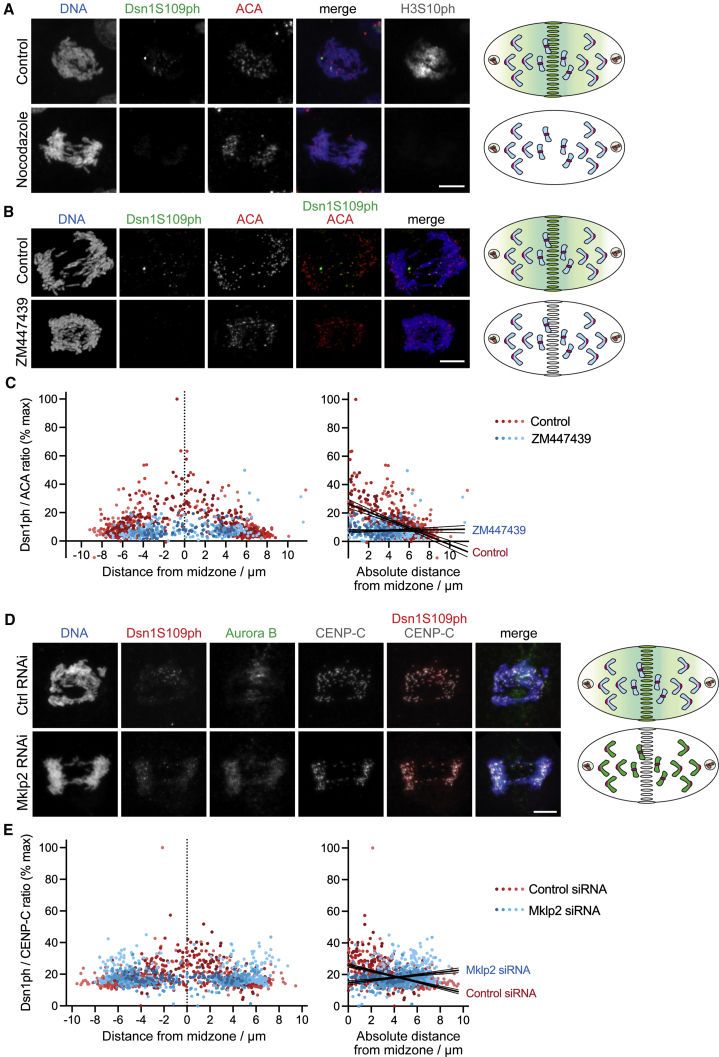
Figure 2.
The gradient of Dsn1S109ph is dependent on central spindle Aurora B
(A) Central spindle depolymerization (see diagram) eliminates the Dsn1S109ph gradient. HeLa cells were enriched in anaphases with lagging chromosomes (see STAR Methods), and nocodazole was added 5 min prior to fixation and staining for DNA (blue), Dsn1S109ph (green), ACA (red), and H3S10ph (gray).
(B) Acute Aurora B inhibition eliminates the Dsn1S109ph gradient. HeLa cells were treated with MPS1-IN-1 and then with 10 μM ZM447439 for 15 min, prior to fixation and staining for DNA (blue), Dsn1S109ph (green), and ACA (red).
(C) Quantification of Dsn1S109ph at individual kinetochores as a function of distance from the midzone in nine control and five ZM447439-treated cells treated as in (B). Dots are shaded as in Figure 1D. Using linear regression, for control cells, slope = −2.9 and is non-zero (p < 0.0001, F test). For ZM447439-treated cells, slope = 0.12 and is not significantly different from zero (p = 0.48). The slopes are significantly different from one another (p < 0.0001, F test).
(D) Mklp2 RNAi prevents transfer of Aurora B to the central spindle (see diagram) and weakens the Dsn1S109ph gradient. Control and Mklp2-depleted HeLa cells were treated with MPS1-IN-1 and stained for DNA (blue), Dsn1S109ph (red), Aurora B (green), and CENP-C (gray).
(E) Quantification of Dsn1S109ph at individual kinetochores as a function of distance from the midzone in eight control and seven Mklp2-depleted cells as in (D). Dots are shaded as in Figure 1D. Using linear regression, for control cells, slope = −1.7 and is non-zero (p < 0.0001, F test). For Mklp2-depleted cells, slope = 0.83 and also is non-zero (p < 0.0001, F test). The slopes are significantly different from one another (p < 0.0001, F test). Confidence intervals (95%) are shown as fine lines.
Scale bars: 5 μm (A, B, and D). See also Figure S2.