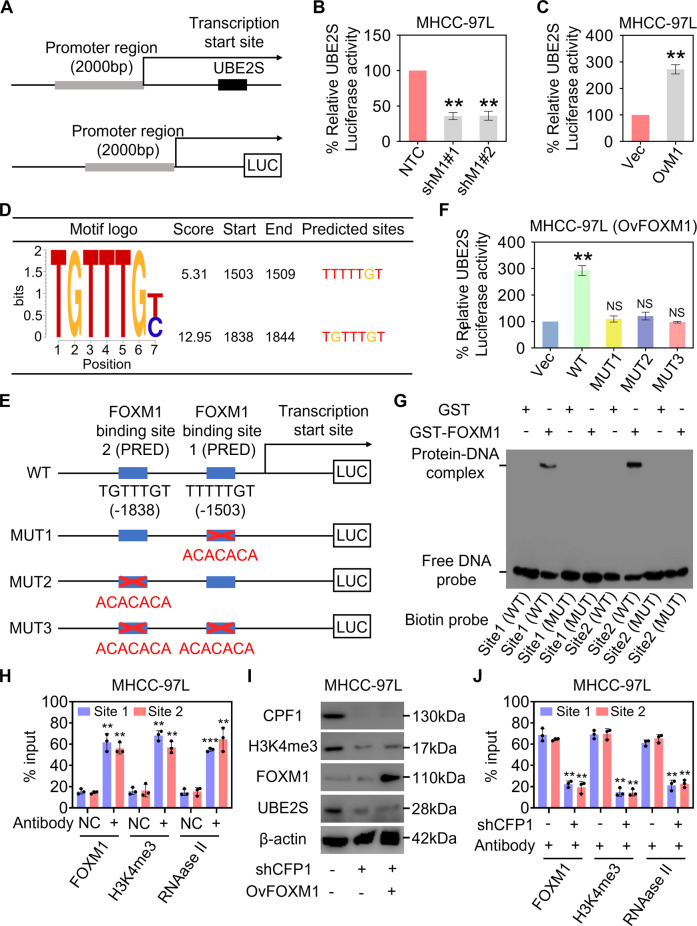
Fig. 4. UBE2S transcription is activated by the binding of FOXM1 to UBE2S promoter.
A Schematic representation of UBE2S promoter. B, C MHCC-97L cells stably silenced FOXM1 (B) or overexpressed FOXM1 (C) were transfected with indicated UBE2S wild-type reporter plasmids, and the luciferase activity was measured, respectively (n = 3). D, E Schematic representation of predicted FOXM1 binding sites (D) and corresponding mutated reporter constructs (E) for UBE2S. F FOXM1-overexpression MHCC-97L cells were transfected with indicated UBE2S wild-type (WT) or mutated reporter plasmids, and the luciferase activity was measured (n = 3). G Biotin-labeled DNA probe from the two predicted FOXM1 binding sites of UBE2S promoter region or corresponding mutant probe was incubated with GST-FOXM1 protein, and the DNA-protein complexes were separated on 6% native polyacrylamide gels. H ChIP-qPCR assays for FOXM1, H3K4me3, and RNA-polymerase II binding sites in the promoter of UBE2S gene (n = 3). I The protein levels of H3K4me3 and UBE2S were investigated in MHCC-97L cells stably silenced CPF1 together with FOXM1 overexpression. J ChIP-qPCR assays for FOXM1, H3K4me3, and RNA-polymerase II binding sites in the promoter of UBE2S gene in CPF1-knockdown MHCC-97L cells (n = 3).