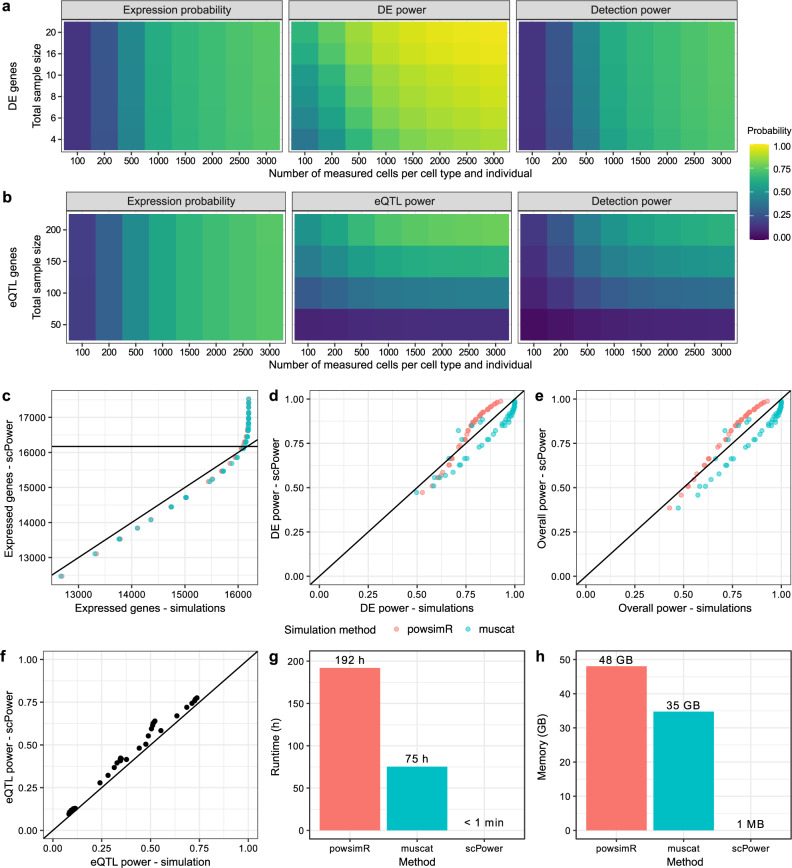
Fig. 3. Expression probability, DE/eQTL power and overall detection power and their validation in simulation studies.
Power estimation using data driven priors for DE genes (a) and eQTL genes (b) dependent on the total sample size and the number of measured cells per cell type. The detection power is the product of the expression probability and the power to detect the genes as DE or eQTL genes, respectively. The fold change for DEGs and the R2 for eQTL genes were taken from published studies, together with the expression rank of the genes. For (a), the Blueprint CLL study with comparison iCLL vs mCLL was used, for (b), the Blueprint T cell study. The expression profile and expression probabilities in a single cell experiment with a specific number of samples and measured cells was estimated using our expression prior, setting the definition for expressed to > 10 counts in more than 50% of the individuals. Multiple testing correction was performed by using FDR adjusted p values for DE power and FWER adjusted p values for eQTL power. The probabilities calculated in (a) were verified by the simulation-based methods powsimR and muscat with each point representing one parameter combination. f The eQTL power of (b) could be replicated with a self-implemented simulation. Runtime (g) and memory requirements (h) were drastically higher in the simulations than for our tool scPower during the evaluations of (c–e), showing the strength of our analytic model.