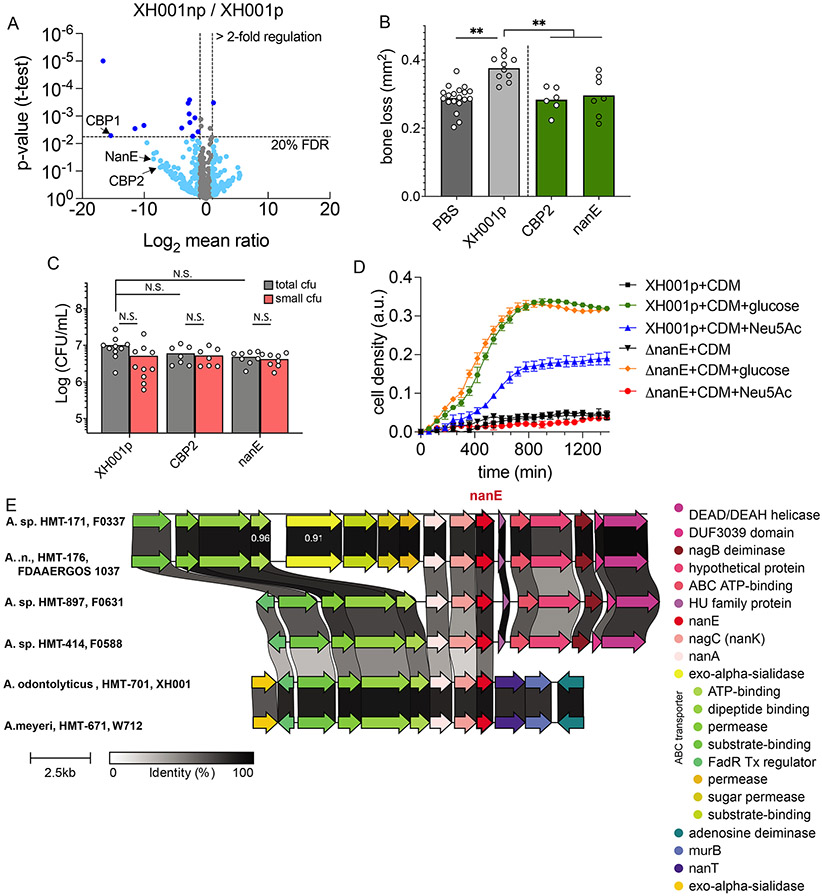
Figure 6.
Modified genes in host bacteria. (A) Comparative mass spectrophotometer analysis of cell wall proteins from XH001p and RC of XH001np shown by volcano plot. The threshold is set for genes that are increased >2-fold (x-axis) and with FDR <20% (y-axis). Dark blue dots are significantly regulated genes by these two criteria, while light blue dots are marginally significantly regulated genes that have higher than 20% FDR. (B) Top genes from panel A were deleted using homologous recombination in XH001p. These mutants were tested in our murine ligature model and the bone loss was measured in each mutant. (C) CFU quantification of panel B mutants on the ligature. (D) Bulk growth curve measurement of XH001p and ΔnanE mutant growing on minimal CDM medium supplemented with buffer, glucose or N-acetylneuraminic acid (Neu5Ac). See also Figure S6. (E) Complete genomes of Actinomyces were downloaded from NCBI RefSeq, aligned, and queried for the nanE gene (red) and its operon. Conserved synteny of nanE operon in three tested Actinomyces host bacteria (XH001, W712, F0337) and three additional taxonomically distance Actinomyces spp. (F0631, F0588, FDAAERGOS 1037). (B and C) Results are shown as mean (bar) ± SD. Each dot indicates an individual ligature (n = 8-10). N.S., not significant; **p < 0.01 by one-way ANOVA followed by Bonferroni post hoc test.