Figure 2. Prolines can function as switches and flexible hinges.
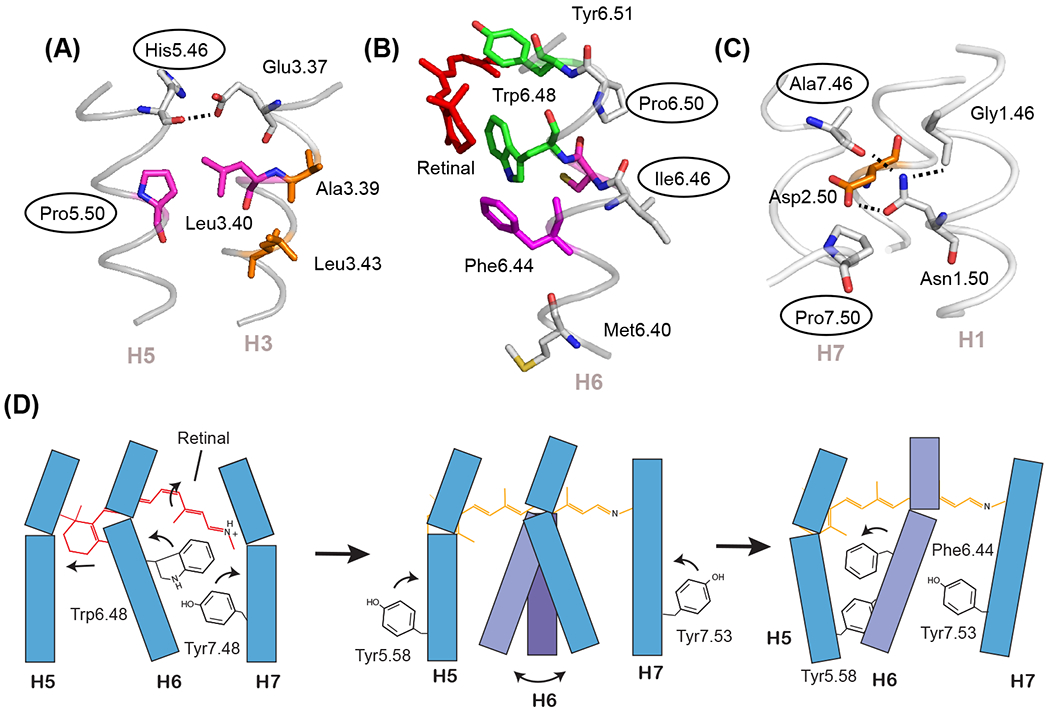
The most conserved residues on TM helices H5, H6, and H7 are prolines that are associated with free carbonyls at positions 5.46, 6.46, and 7.46. (A) In the inactive state of rhodopsin (PDB-ID 1GZM), the His5.46 backbone carbonyl hydrogen-bonds with Glu3.37 on H3. Upon activation, there is a switch in hydrogen-bonding partners to form direct side chain His5.46 - Glu3.37 contacts. The side chain at residue 5.46 is subfamily specific (e.g. in the adrenergic receptors this residue is part of a conserved SSxxS motif that hydrogen-bonds to hydroxyl groups on the catechol ring of agonist ligands). (B) The free C=O group at position 6.46 associated with Pro6.50 is oriented toward the surrounding lipids and consequently is not in a position to form interhelical hydrogen-bonds. This region is predisposed to function as a flexible hinge. (C) The free C=O group at position 7.46 is conserved as a small residue (typically Ala or Ser) in class A GPCRs and serves to orient the side chain of Asn1.50. (D) Schematic showing key steps in rhodopsin activation. Helices H1-H4 (not shown) form a structural scaffold on which helices H5-H7 pack. Retinal isomerization (left panel) releases packing constraints on Trp6.48 which disrupts the Trp6.48-Tyr7.48 interaction and imparts flexibility at the H6 proline hinge (middle panel). An outward orientation of H6 is stabilized by the rotation and repacking of three aromatic and hydrophobic residues. Key residues are Phe6.44, Tyr5.58, and Tyr7.53 (right panel).
