Figure 6. Hydrophobic residue packing in active conformations of switch 1 and 2.
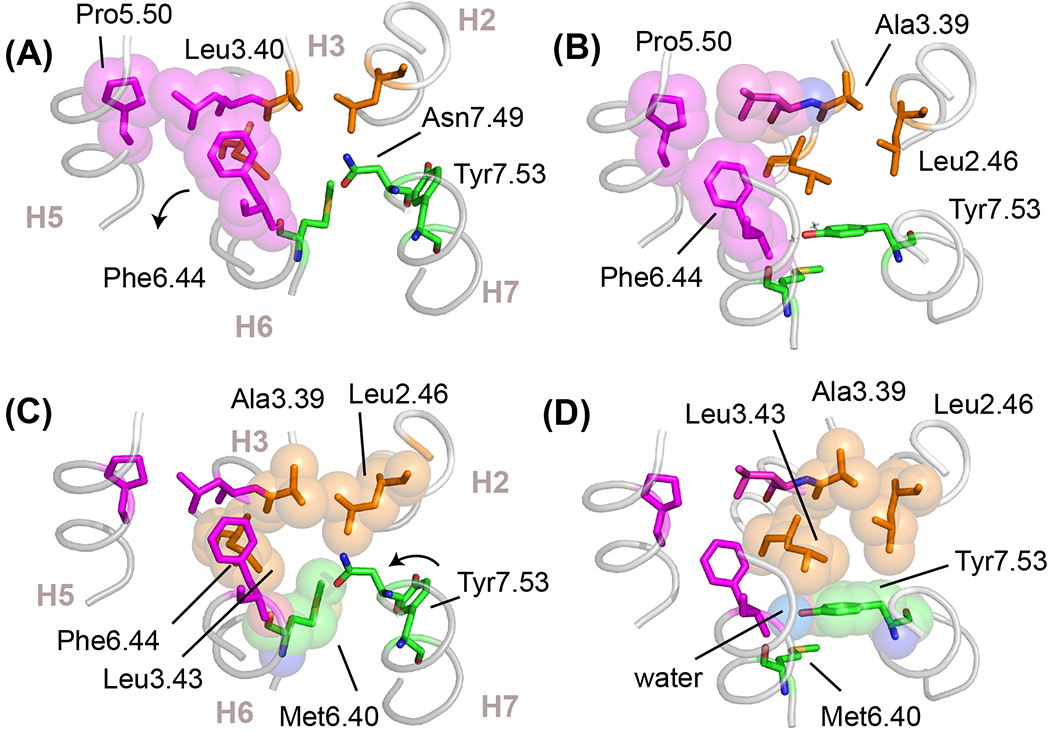
(A,C) Structure of the inactive state of rhodopsin (PDB-ID 1GZM) highlighting hydrophobic packing interactions in switch 1 and switch 2, shown in magenta and orange spheres, respectively. In the inactive state, Phe6.44 in switch 1 stacks with Trp6.48 (see Figure 2B) and Met6.40 in switch 2 packs against Leu2.46, Leu3.43 and Asn7.49. Position 6.40 varies in different subfamilies, but is often found (>50%) as a valine or isoleucine. In rhodopsin, this hydrophobic interaction stabilizes the inactive conformation [59]. (B,D) Structure of the active state of rhodopsin (PDB-ID 3PQR) showing the changes in hydrophobic packing interactions. In switch 1, Phe6.44 rotates to form a packing contact with Pro5.50. In switch 2, the outward rotation of H6 disrupts the packing contact formed between Met6.40 and the other residues in switch 2. The conserved Tyr7.53 of the NPxxY motif swings up and packs into the space vacated by Met6.40 and packs against Leu2.46 and Leu3.43. The Cζ-OH of Tyr7.53 forms a water-mediated hydrogen bond with a conserved water molecule that bridges the backbone C=O residues of Met6.40 and Leu3.43. Water is shown as a blue van-der Waals sphere in panel (D).
