Figure 4. Structure and dynamics of the mammalian outer dynein arm (ODA) and ODA docking complex (ODA-DC).
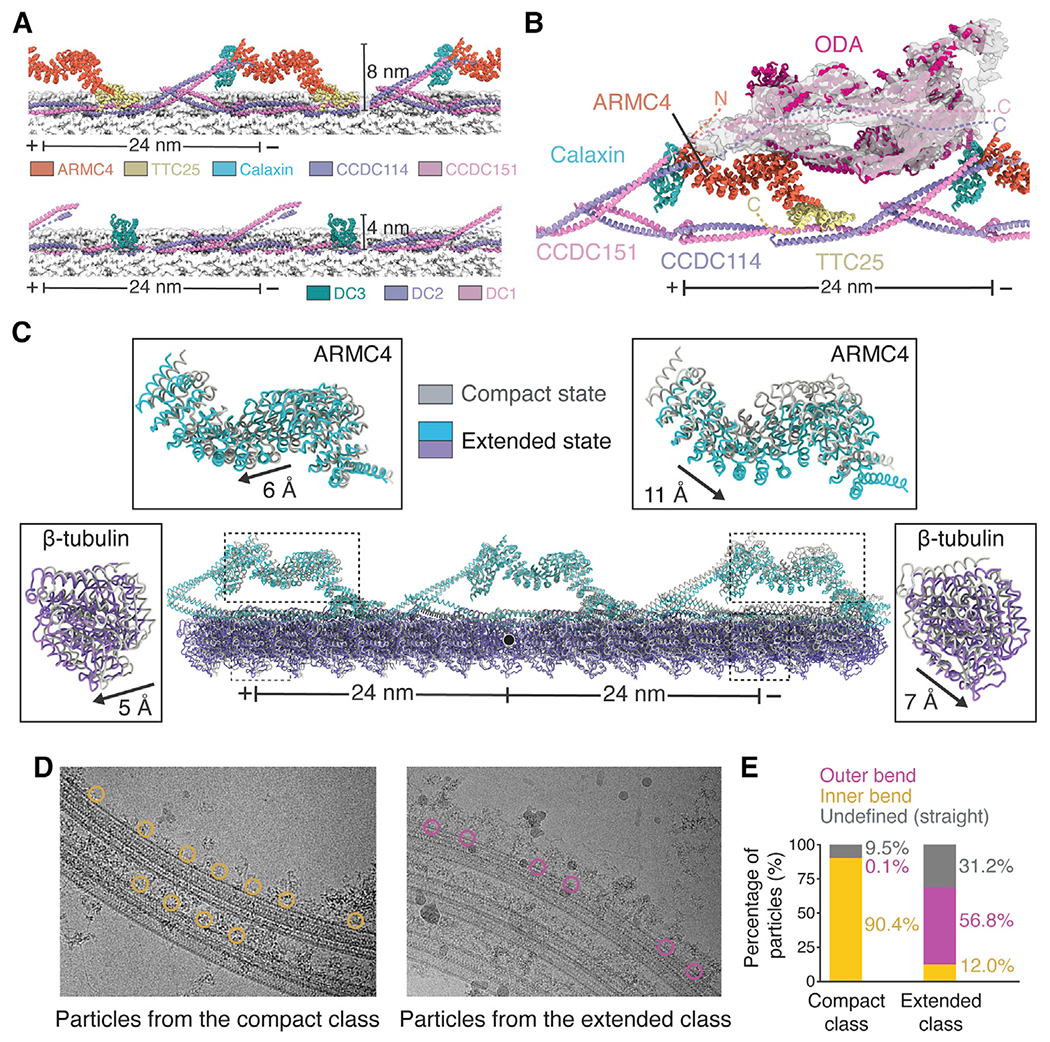
(A) Models of bovine (top) and Chlamydomonas (bottom) ODA-DCs. Tubulin is shown in surface representation.
(B) Model of the Chlamydomonas ODA (PDB: 7KZM) docked into the cryo-EM map of the bovine ODA. Dashed lines represent the predicted locations of the termini of ODA-DC subunits when ODA is bound.
(C) Analysis of tubulin lattice spacing reveals an extended and compact conformation with different ODA-DC conformations. The two models are superposed on the central tubulin (marked with a black circle). The displacement of ARMC4 and β-tubulin between classes was calculated using the mass center of the molecules.
(D) Particles from the compact and extended classes mapped back onto the micrographs.
(E) Quantification of the particle locations observed in (D). Only micrographs with 8 or more particles were analyzed. Compact class, n = 1,017; extended class, n = 1,201.
