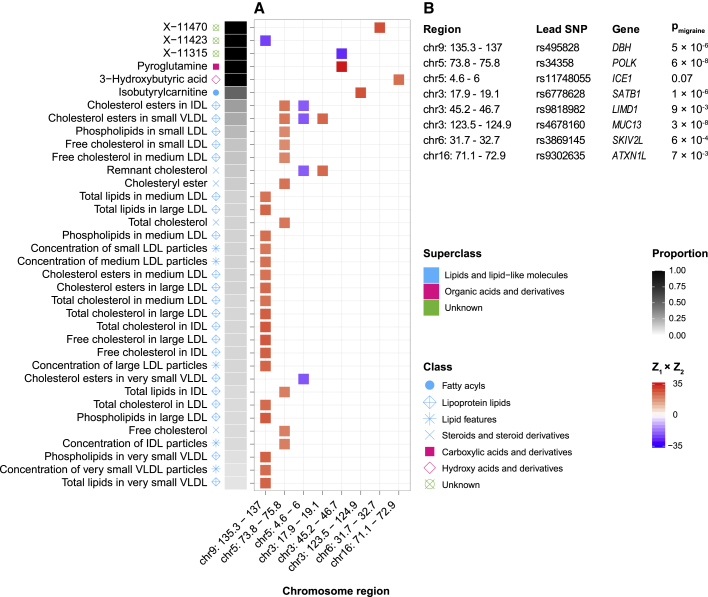
Figure 3.
Pleiotropic-associated loci influencing migraine and blood metabolome (genetic overlap at the loci level)
(A) GWAS-PW identified eight independent pleiotropic loci across five chromosomes, influencing migraine risk and 36 metabolic traits levels (out of 316 tested metabolic traits) via a SNP within each locus. The concordance (red color) or discordance (blue color) of each locus is defined by the Z score of the lead migraine SNP from the migraine GWAS and from each metabolic trait GWAS (Z1 × Z2). The proportion of the independent loci influencing the metabolic trait also influencing migraine is shown in gray to black.
(B) For the identified pleiotropic loci, lead SNPs from the migraine GWAS (lead migraine SNPs) are shown. We used MAGMA results (pmigraine) to identify lead migraine genes in the identified pleiotropic loci. Different colors and shapes represent superclasses and classes, defined by the HMDB, respectively.