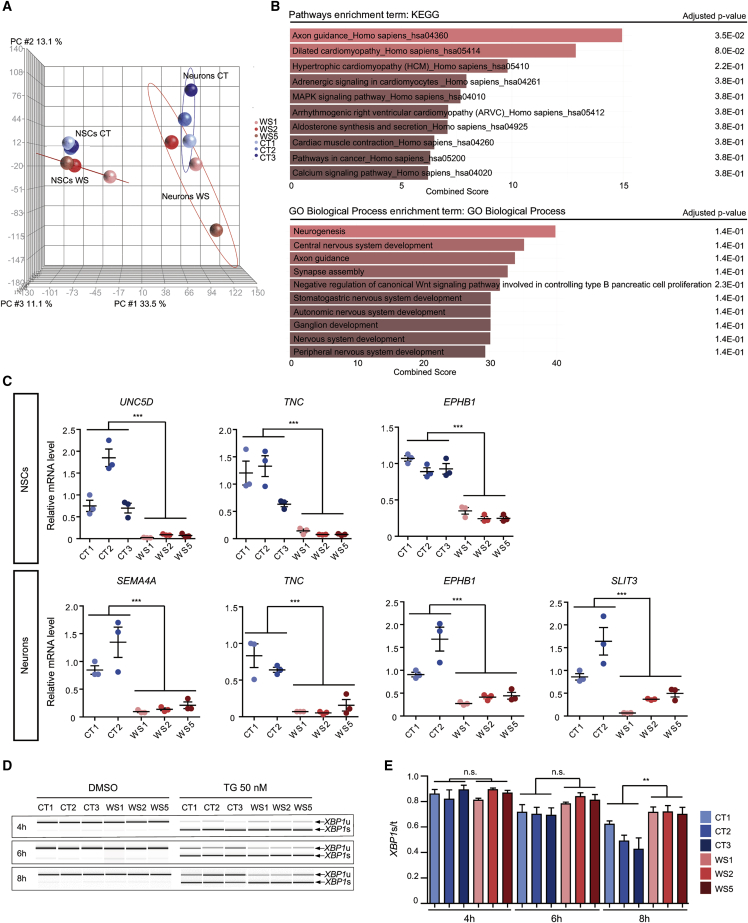
Figure 4.
Global gene expression comparison between CT and WS NSCs and neurons
(A and B) Transcriptome analysis with AmpliSeq. (A) Principal-component analysis (PCA) between CT and WS NSCs and neurons. (B) KEGG pathways (upper panel) and gene ontology (lower panel) enrichment analysis in neurons via Enrichr tool.
(C) Quantitative RT-PCR analysis of DEGs mRNA levels in CT and WS NSCs and neurons. Data are expressed relative to that of CT1 cells and normalized to 18S rRNA expression. Data are presented as mean ± SEM (n = 3 biologically independent differentiations; ∗∗∗p < 0.001; Student’s t test).
(D and E) ER stress induction in CT and WS NSCs after 4, 6, and 8 h of 50 nM thapsigargin (TG) treatment. (D) Analysis of the expression of spliced (XBP1s) and unspliced (XBP1u) XBP1. (E) Quantification of XBP1s expression normalized to total XBP1 (XBP1t). Data are presented as mean ± SEM (n = 3 biologically independent experiments; ∗p < 0.05; one-way ANOVA with Dunnett’s post hoc test).