Extended Data Fig. 5. Related to Figure 6.
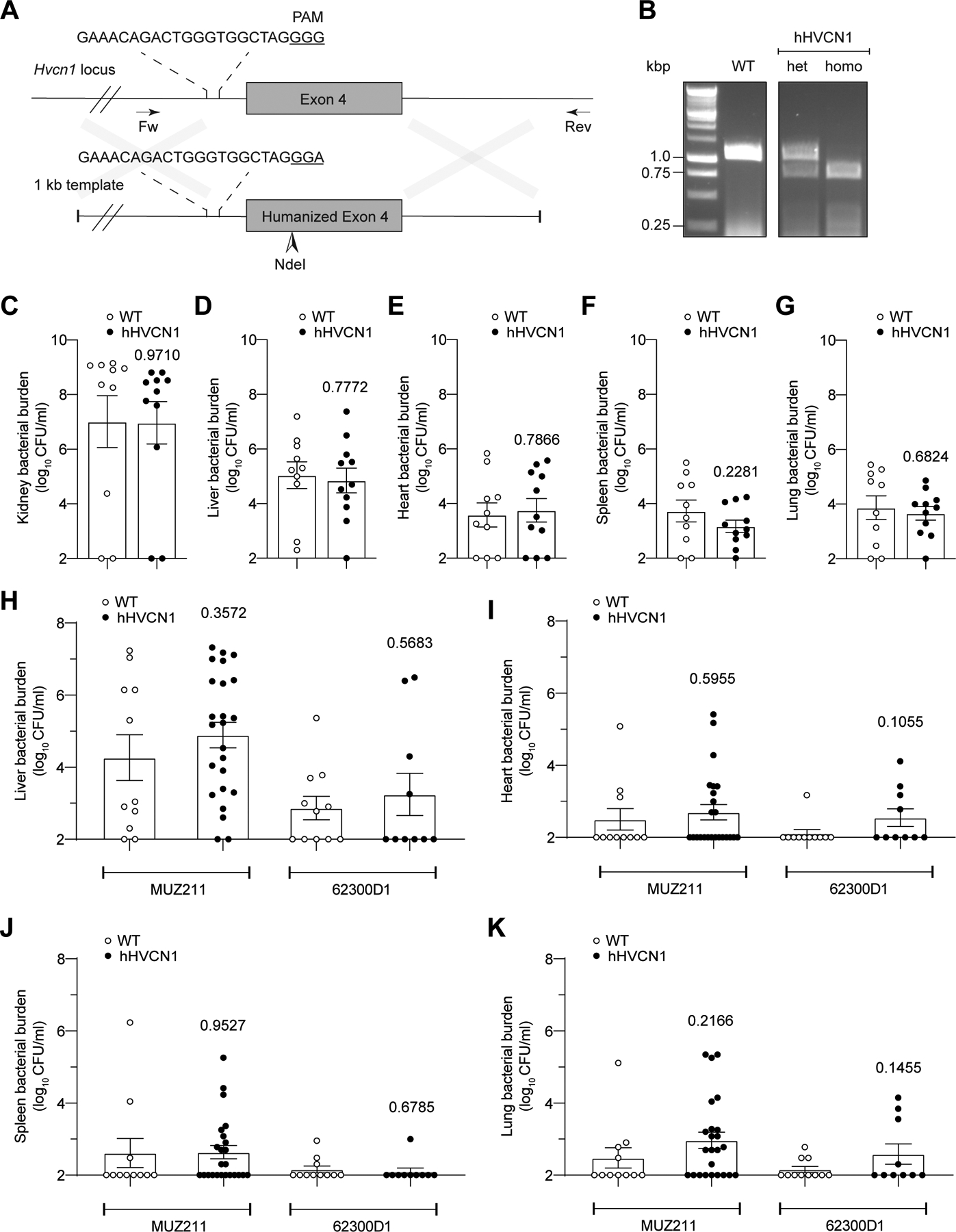
A: Schematic representation of murine Hvcn1 locus and DNA template used to humanize exon 4. B: Genotyping strategy using genomic DNA isolated from wild type (WT), heterozygous (het), and homozygous (homo) hHVCN1 mice using primers VJT2065 and VJT2069. Images are representative of multiple independent experiments as routinely performed for hHVCN1 mouse genotyping. C-G: CFUs in the kidneys (C), livers (D), hearts (E), spleens (F), and lungs (G) collected from WT and hHVCN1 mice infected intravenously with 1×107 CFU of lukAB-deficient USA300 strain LAC. Data from 11 WT and 10 hHVCN1 mice are represented as mean values ±SEM. Statistical significance was determined by t-test (two-tailed), numbers above bars indicate P values. H-K: CFUs in the livers (H), hearts (I), spleens (J), and lungs (K) collected from WT and hHVCN1 mice infected intravenously with 5–10×107 CFU CC30 S. aureus MUZ211 (CFU obtained from 11 WT and 24 hHVCN1 mice) and 62300D1 (CFU obtained from 11 WT and 10 hHVCN1 mice). Data for each isolate are from mice infected over three independent experiments and is represented as mean values ±SEM. Statistical significance was determined by t-test (two-tailed), numbers above bars indicate P values.
