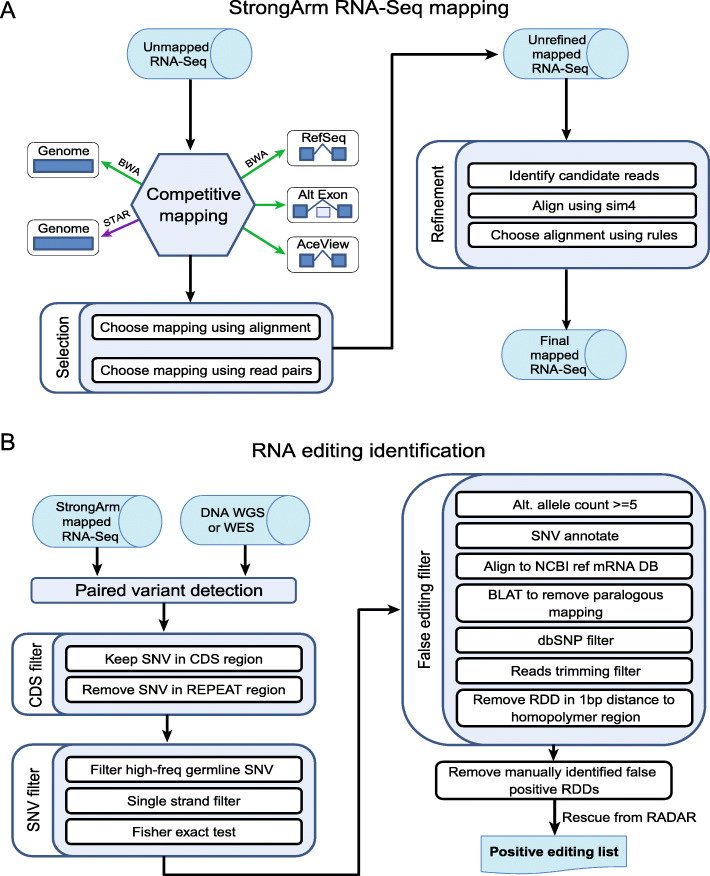
Fig. 1.
The StrongArm RNA-Seq mapping and RNA editing detection pipelines. A Schematic workflow of StrongArm RNA-seq mapping pipeline. The pipeline starts with competitive mapping of 5 different combinations of mapper and database, followed by further local refinement. B RNA editing identification pipeline. RNA-Seq BAM files are aligned with StrongArm as shown in (A), and germline and somatic DNA variants are also called from the same patient using WGS or WES of matched tumor and germline DNA. The pipeline searches for RNA-specific (RNA editing) variants in coding (CDS) regions by comparing RNA-Seq reads to DNA-Seq. A series of false editing filters is then employed to remove RNA editing artifacts, followed by manual review of the BAM alignment. The RNA editing candidates are then used to evaluate the editing levels cross the whole cohort