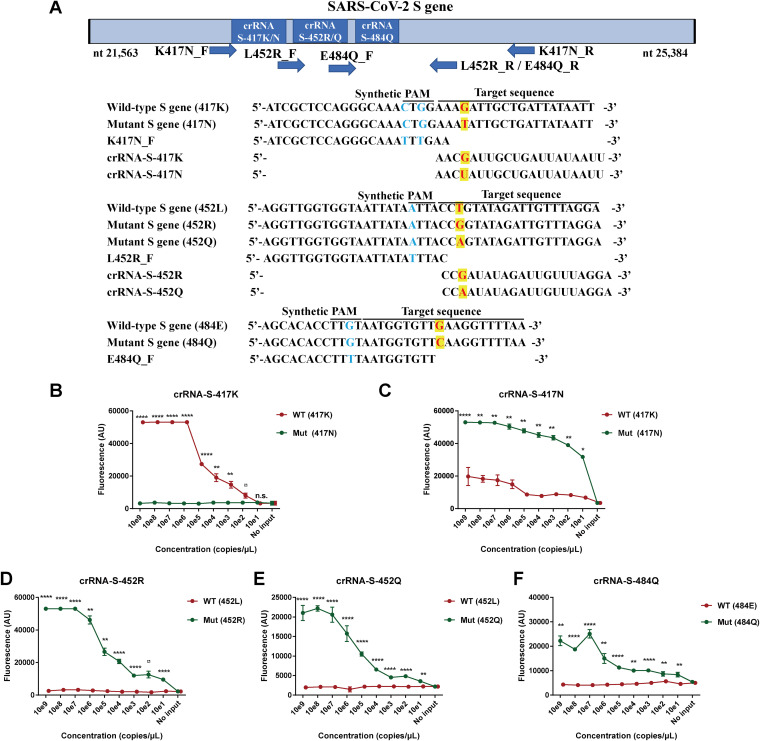
FIG 6.
Strategy of adding protospacer adjacent motif (PAM) near mutation sites by using PCR primers to promote CRISPR/Cas12a-mediated detection of SARS-CoV-2 mutants. (A) Schematic of the SARS-CoV-2 spike (S) genome, forward primer with PAM sequence, and the sequences and locations of the corresponding guide RNAs crRNA-S-417K, crRNA-S-417N, crRNA-S-452R, crRNA-S-452Q, and crRNA-S-484Q for SARS-CoV-2 mutants 417N, 452R/Q, and 484Q, respectively. Different concentrations of SARS-CoV-2 DNA were used as the template and amplified by PCR with the aforementioned forward primers followed by detection of CRISPR-Cas12a-mediated assay to detect the single nucleotide mutation of K417N (B and C), L452R (D), L452Q (E), and E484Q (F), respectively. The fluorescence values represent the mean ± standard deviation (SD) of 3 replicates. Two-tailed Student's t test was used to analyze the difference between wild type and mutant template. n.s., not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001; WT, wild type; Mut, mutant; no input, negative control with no plasmid DNA.