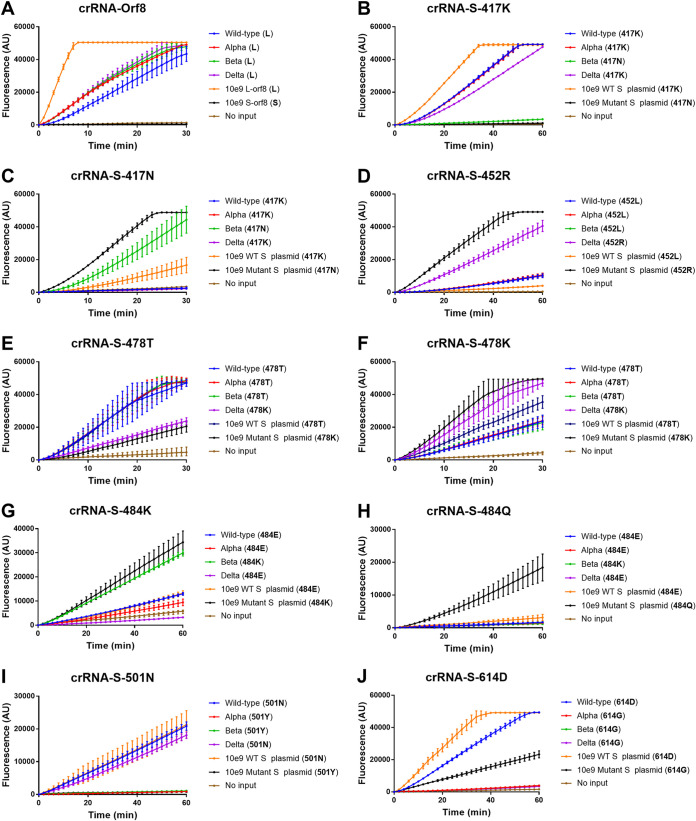
FIG 8.
CRISPR-Cas12a-based detection of SARS-CoV-2 variants. Viral RNA templates were extracted from SARS-CoV-2 wild-type strain as well as alpha, beta, and delta SARS-CoV-2 variants of concern and amplified by RT-PCR followed by CRISPR-Cas12a-mediated detection by using crRNA-Orf8 (A), crRNA-S-417K (B), crRNA-S-417N (C), crRNA-S-452R (D), crRNA-S-487T (E), crRNA-S-487K (F), crRNA-S-484K (G), crRNA-S-484Q (H), crRNA-S-501N (I) and crRNA-S-614D (J) for the corresponding mutations. The position and name of the amino acid for the templates are indicated in the brackets. The plasmid DNA for S-lineage Orf8 (S-Orf8; 109 copies/μl) and L lineage Orf8 (L-Orf8, 109 copies/μl) gene is used as control in panel A. The plasmid DNA for wild-type (WT, 109 copies/μl) and mutant spike (S, 109 copies/μl) genes was used as control in the remaining panel B to J. The fluorescence was measured at different time points and presented as the mean ± standard deviation (SD) from 3 replicates. No input, negative control with no plasmid DNA.