FIG 2.
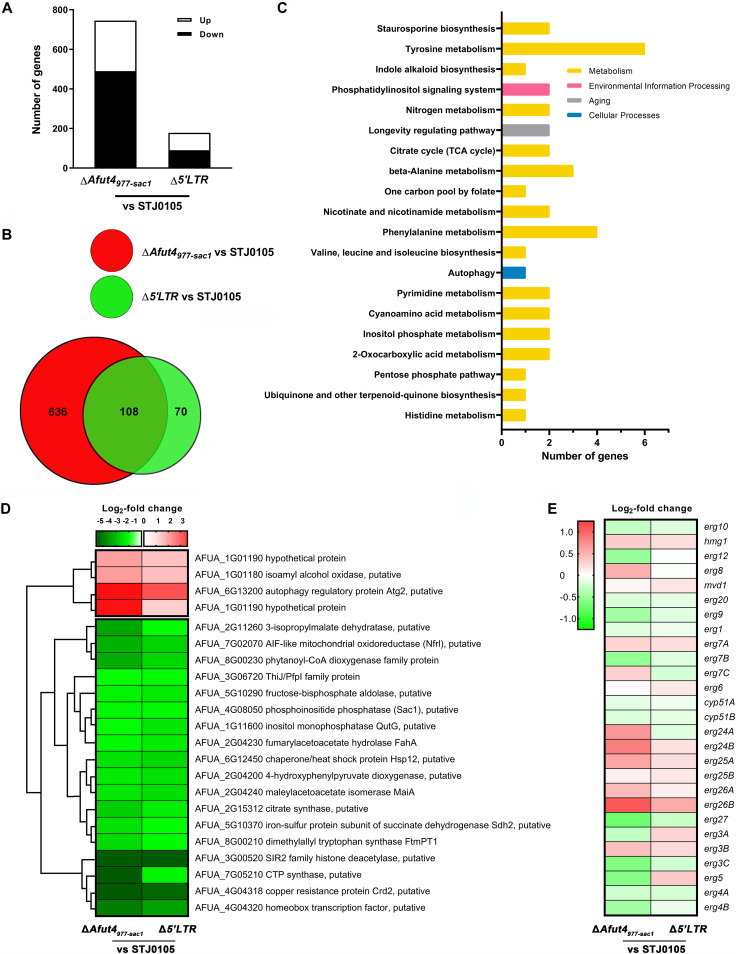
RNA sequencing analysis of STJ0105, ΔAfut4977-sac1, and Δ5'LTR. (A) A summary of the differently expressed genes (DEGs) in ΔAfut4977-sac1 relative to STJ0105 (ΔAfut4977-sac1 vs STJ0105) and Δ5'LTR relative to STJ0105 (Δ5'LTR vs STJ0105) (|log2 fold change ≥ 1|; Q < 0.05). (B) Venn diagram showing the differentially and commonly shared DEGs (|log2 fold change ≥ 1|; Q < 0.05) in ΔAfut4977-sac1 versus STJ0105 and Δ5'LTR versus STJ0105. (C) The top 20 enriched pathways of DEGs (|log2 fold change| ≥ 1; Q < 0.05) from the 108 shared DEGs in both ΔAfut4977-sac1 versus STJ0105 and Δ5'LTR versus STJ0105. According to KEGG annotations and classifications, the 58 significant DEGs were classified into different biological pathways. KEGG enrichment analysis was carried out by using the phyper function in R software. The top 20 enriched pathways were screened from all the biological pathways. (D) Hierarchical cluster analysis of the log2 fold change of 22 significant DEGs in the top 20 enriched pathways. (E) Log2 fold change of genes involved in ergosterol biosynthesis in ΔAfut4977-sac1 and Δ5'LTR versus STJ0105.