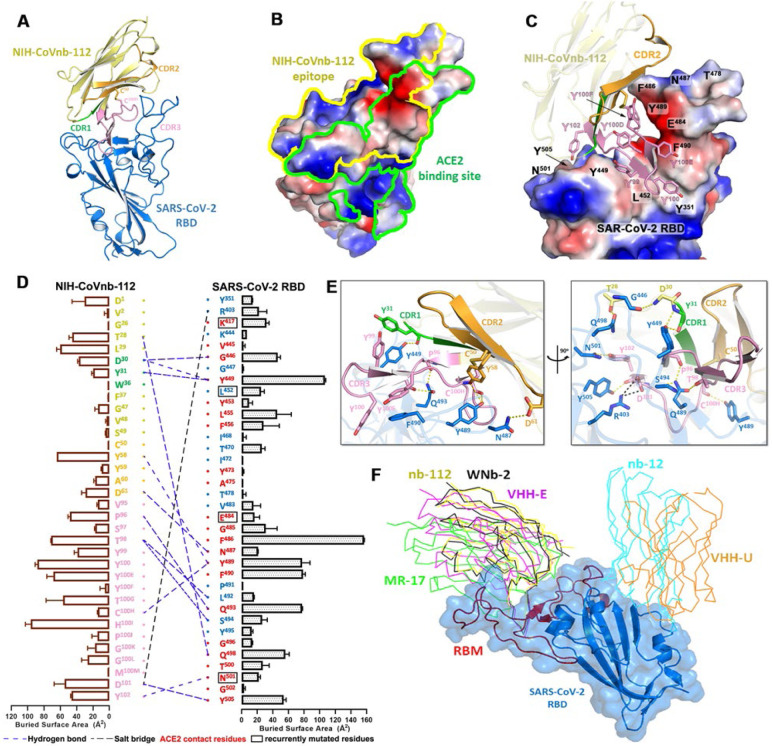
Figure 1. Molecular basis of SARS-CoV-2 spike recognition by NIH-CoVnb-112.
(a) Crystal structure of NIH-CoVnb-112:SARS-CoV-2 RBD complex. The framework of NIH-CoVnb-112 is shown as yellow ribbons with the CDR1, CDR2, CDR3 colored green, orange and pink, respectively. The SARS-CoV-2 RBD is colored blue. (b) Comparison of the RBD-contact footprint of NIH-CoVnb-112 and ACE2. The contact surface area is indicated by yellow and green lines for NIH-CoVnb-112 and ACE2, respectively, over the molecular surface of RBD. The electrostatic potential is displayed over the RBD surface and colored for negative (red), positive (blue), and neutral (white) electrostatic potential, respectively. (c) Enlarged view into the NIH-CoVnb-112:RBD interface. NIH-CoVnb-112 is shown as a ribbon with the same color scheme as (a) and the SARS-CoV-2 RBD is displayed as an electrostatic potential surface. (d) The interaction network at the NIH-CoVnb-112:RBD interface. The individual nanobody:RBD contacts are shown as lines with the diagram of buried surface area for individual interface residue shown on the sides. Nanobody residues in the framework and CDRs are colored as in (a) with interactions defined by a 5-Å distance criterion cutoff shown as lines. The Kabat19 scheme was used to number nanobody amino acid residues, with unique insertion residues indicated by letter (e.g., 52a, 52b, 52c). Salt bridges and H-bonds (bond length less than 3.5 Å) are shown as black and blue dashed lines, respectively. The ACE2 contact residues within the nanobody epitope are highlighted in red or otherwise in blue. The recurrently mutated RBD residues in SARS-CoV-2 variants are marked with black boxes. (e) Orthogonal views of the interaction network between NIH-CoVnb-112 and RBD. CDR residues are colored as in (a) and the RBD residues are colored in blue. Hydrogen bonds and salt bridges are denoted as yellow and black broken lines, respectively. (f) Comparison of the mode of RBD recognition by NIH-CoVnb-112 to other RBD-specific nanobodies. The NIH-CoVnb-112-RBD complex was superimposed (based on the RBD) to five published RBD complex structures of RBD-specific nanobodies, including WNb-2 (PDB: 7LDJ), VHH-E (7KN5), MR17 (7CAN), Nb12 (7MY3), and VHH-U (7KN5). The SARS-CoV-2 RBD is shown in a semi-transparent blue surface with the receptor-binding motif (RBM) highlighted in red. Other nanobodies are shown with colored lines as labeled.