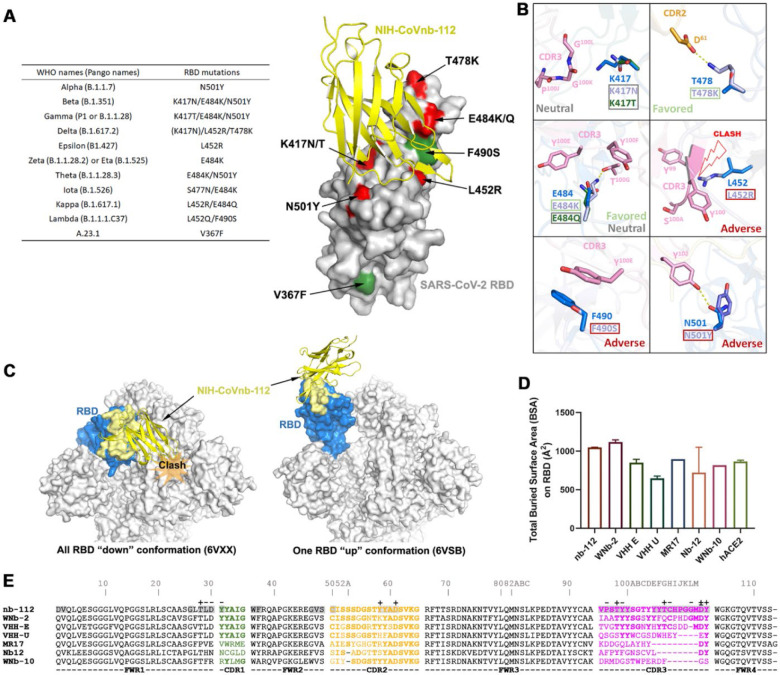
Figure 2. Crystal structure of NIH-CoVnb-112 in complex with SARS-CoV-2 RBD and effects of RBD mutations.
(a) Diagram of RBD mutations occurred in the circulating SARS-CoV-2 variants. The recurrent RBD mutations within other SARS-CoV-2 variants are colored in red with unique mutations in green. (b) Composite model of RBD escape mutations on or around the epitope of NIH-CoVnb-112. The nanobody and prototype RBD are colored as in Fig. 1. The selected RBD mutations with side chain depicted only are shown in indicated colors. (c) RBD-based superimposition of the NIH-CoVnb-112-RBD complexes with SARS-CoV-2 spike in a closed (PDB: 6VXX)46 or one-RBD-up state (PDB: 6VSB).47 The nanobody is shown as yellow ribbons and the RBD domain is colored in blue with the nanobody epitope highlighted in yellow. The potential clashes of nanobody with the adjacent RBD of the closed spike trimer is shown in the left panel. (d) Comparisons between the RBD total buried surface areas (BSAs) of NIH-CoVnb-112, ACE2 and six published RBD-directed nanobodies. (e) Sequence alignments of NIH-CoVnb-112 with six structurally characterized RBD-binding nanobodies. The CDR sequences are colored as indicated in Fig 1a. The buried surface residues (BSA > 0 Å) as calculated by PISA are shaded in grey. Contact residues involved in salt-bridges or H-bonds to the RBD are marked above the sequence with (+) for the side chain and (−) for the main chain. The Kabat19 scheme was used to number nanobody amino acid residues, with unique insertion residues indicated by letter.