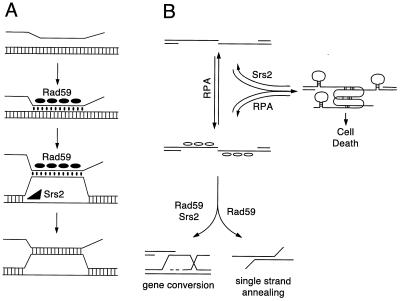
FIG. 9.
Models demonstrating how nonhomologous tails might impede DSB-induced gene conversion in rad59 mutants. (A) During DSB-induced gene conversion, a single-stranded DNA intermediate searches duplex DNA for a homologous sequence and initially forms a paranemic joint that is converted to a plectonemic joint. The presence of nonhomologous sequences at the end of the invading strand destabilizes the paranemic joint or impedes the conversion to a plectonemic joint (43). Rad59p (closed ellipses) facilitates the process by annealing the strands and stabilizing the joint. A stable paranemic joint in turn provides the opportunity for the tail to be removed or for a topoisomerase to act. In the absence of Rad59p, longer tails pose a greater impediment to the formation of a plectonemic joint. This is consistent with a previous interpretation of the srs2 mutation, which behaves similarly in the DSB-induced gene conversion assay (38). Srs2p (black triangle) is proposed to stabilize the same structure by unwinding the donor duplex, allowing easier formation of a paranemic joint and, later, a plectonemic joint. (B) When nonhomologous tails become long, they may impede SSA or DSB-induced gene conversion by forming secondary structures in a process mediated by the annealing of microhomologous sequences. The single-stranded DNA binding protein RPA (open ellipses) may reduce secondary structures by forming complexes with single-stranded DNA. These complexes likely lie in equilibrium with the unbound state as the single-stranded tail searches for homologous sequence. As such, the secondary structures remain competitors to successful SSA or gene conversion. A mutation such as rad59 or srs2 that cripples SSA or gene conversion will shift the equilibrium toward the secondary structures, eventually leading to cell death. Substrates with short tails or no tails do not have this competing pathway, and hence it only reveals itself with structures with long tails in a mutant background. Mutations in srs2 decrease SSA to a lesser extent than do rad59 mutations, which is consistent with the likelihood that a helicase is not required for annealing of complementary strands. Srs2p may promote SSA indirectly by unwinding microhomology-based secondary structures or by aiding in the homology search process by unwinding improperly annealed single-stranded tails.