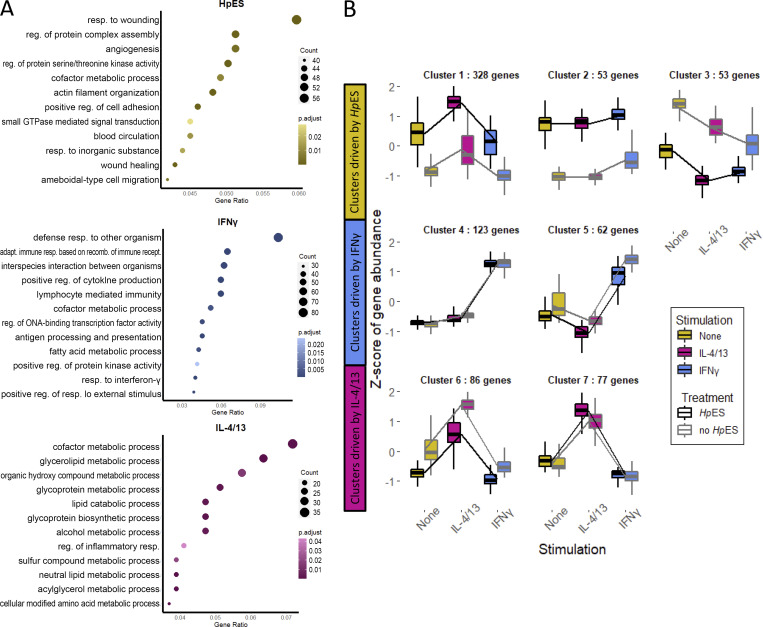
Figure S1.
GO terms for DEGs. (A) HpES and cytokine treatment of small intestinal organoids. Top 12 GO terms, by gene ratio, for DEGs from HpES, IL-4/13, and IFN-γ individual treatments. Data are based on analyses of four biological replicates, composed of a total of 24 samples analyzed in parallel by RNA-seq. DEGs were selected as those with a P-adjusted value of <0.01. The enrichGO function from ClusterProfiler (Yu et al., 2012) was used to identify enriched GO terms, followed by ReViGO (Supek et al., 2011) to remove redundant GO terms. The top 12 GO terms were then selected using gene ratio (number of genes associated with GO term in list/total number of genes in list). (B) Responses of identified gene clusters to HpES, IL-4/13, and IFN-γ. Gene sets were split based on treatment (± HpES) and stimulation (IL-4/13, IFN-γ, or none). Clusters were identified using degPatterns, a part of the DEGreport package (Pantano et al., 2020). Adapt., adaptive; GTPase, guanosine triphosphatase; rec., reecombinant; reg., regulation; resp., response.