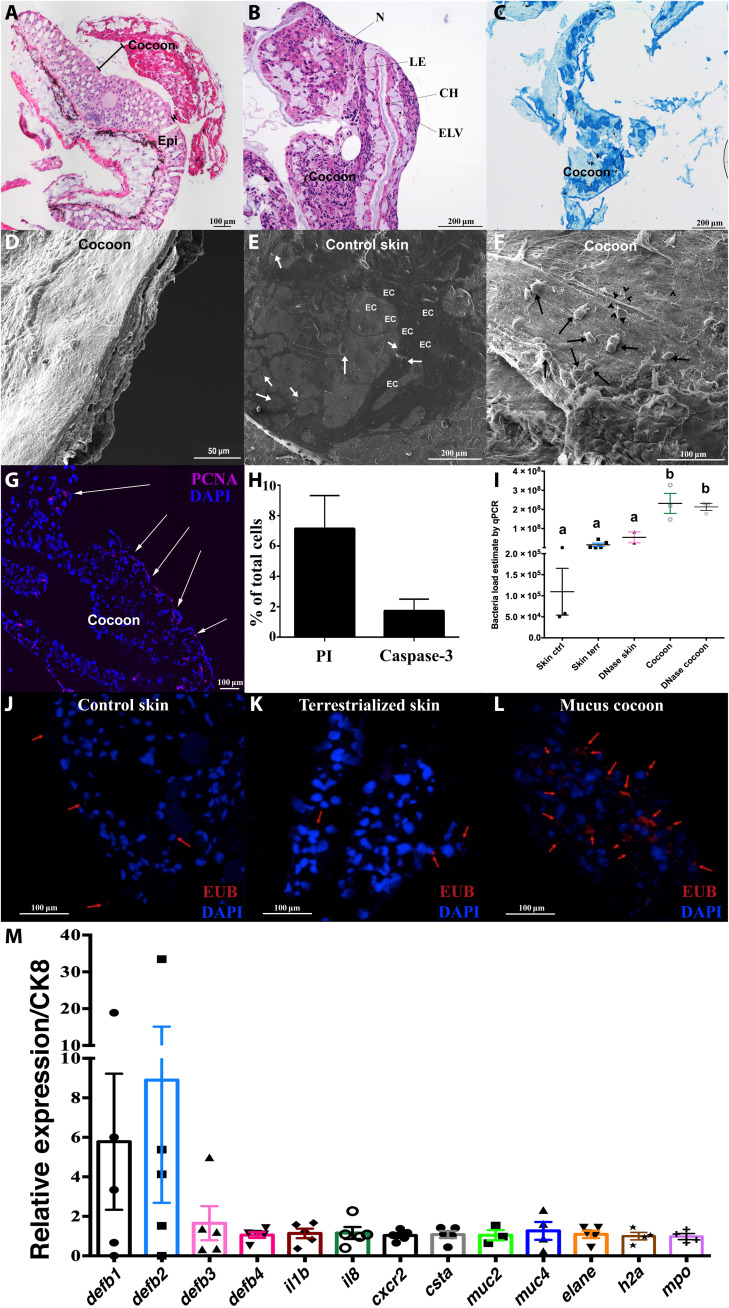
Fig. 3. The lungfish mucus cocoon is a living cellular structure that traps bacteria.
(A) H&E staining of lungfish cocoon (large image composite). (B) Magnified view of an H&E-stained cocoon. N, nest of cells; CH, channels; IC, intercommunicating channels; LE, loss of eosinophilia of nest cells; ELV, endothelium lined vessels. (C) Periodic acid–Schiff staining of lungfish cocoon showing goblet cells (dark blue) and mucus but no positive staining in the channels (n = 3). (D) SEM image of lungfish cocoon cross section showing uneven cellular layers. (E) Coronal SEM image of control lungfish skin showing polygonal epithelial cell (EC) and goblet cell openings (white arrows). (F) Coronal SEM image of cocoon showing lungfish cells (black arrows) and bacteria (black arrow heads). (G) PCNA staining (magenta) of a cocoon cryosection showing proliferating cells at the edge (white arrows). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). (H) Percent PI+ and activated caspase-3+ cells in cocoon single-cell suspensions (n = 3). (I) Relative quantification of bacterial loads by qPCR in control skin, terrestrialized skin, and mucus cocoon (n = 3 to 4). Different letters indicate statistically significant differences [one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test]. (J to L) Maximum projection of confocal fluorescence images of fluorescence in situ hybridization (FISH) using EUB338 oligoprobe (red) showing bacteria (red arrows) in control skin, terrestrialized skin, and mucus cocoon (n = 3). (M) Gene expression of antimicrobial peptide genes (defb1 to defb4), proinflammatory cytokines (il1b and il8), cystatin A (csta), mucins (muc2 and muc4), granulocyte markers (cxcr2, mpo, and elane), and h2a in the lungfish cocoon (n = 5) by RT-qPCR. Cytokeratin 8 (ck8) was used as house-keeping gene.