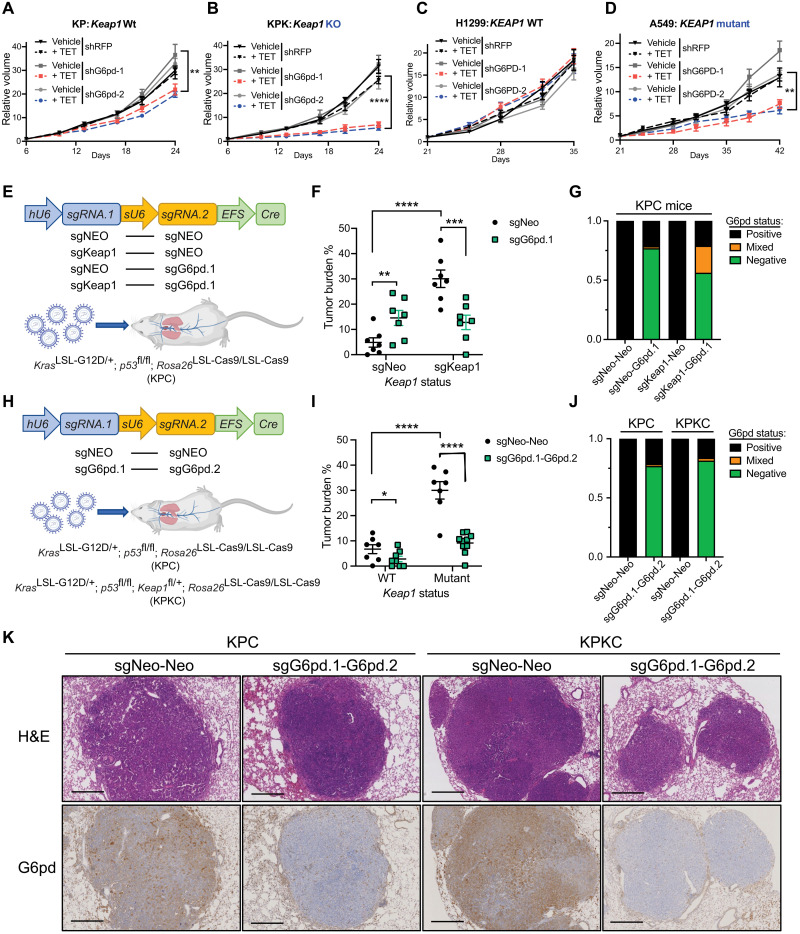
Fig. 4. G6PD is required for KEAP1 mutant LUAD.
(A and B) Relative tumor growth of subcutaneous KP (A) and KPK (B) tumors with inducible shG6pd in animals receiving TET or control diet. TET stands for doxycycline diet activating shRNA expression. Diet was changed at day 6. Data were normalized to first measurement at day 6 (n = 7). (C and D) Relative tumor growth of subcutaneous H1299 (KEAP1 WT) (C) and A549 (KEAP1 mutant) (D) tumors with inducible shG6PD in animals receiving TET or control diet. Diet is changed at day 21. Data were normalized to first measurement at day 21 (n = 8 for H1299 and n = 6 for A549). (E) Schematic figure of KPC mice intratracheally infected with pUSEC lentiviruses containing double sgRNAs targeting G6pd, Keap1, or Neo (Control). (F) Quantification of tumor burden (tumor area/total lung area) in KPC mice after infection with pUSEC lentiviruses (n = 8 for sgNeo-Neo and sgKeap1-Neo and n = 9 for sgNeo-G6pd.1 and sgKeap1-G6pd.1). (G) G6pd status analysis based on IHC staining of lung bearing tumors (n = 4). (H) Schematic figure of KPC or KPKC mice intratracheally infected with pUSEC lentiviruses containing double sgRNAs targeting Neo-Neo and G6pd.1-G6pd.2. (I) Quantification of tumor burden of mice infected with pUSEC lentiviruses (n = 7 for Keap1 WT sgNeo-Neo and sgG6pd.1-G6pd.2, n = 8 for Keap1 mutant sgNeo-Neo, and n = 10 for Keap1 mutant sgG6pd.1-G6pd.2). (J) G6pd status analysis based on IHC staining (n = 3). (K) Representative IHC staining of serial sections from lung tumors of KPC and KPKC mice 16 weeks after infection with pUSEC lentiviruses against Neo-Neo and G6pd.1-G6pd.2. First panels, hematoxylin and eosin (H&E) staining analyses; second panels, G6pd IHC analyses. Note that Keap1 mutant group has higher staining of G6pd, proving G6pd is Keap1/Nrf2 substrate. Scale bars, 500 μm. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Detailed statistics analysis of (A) to (D) is presented in table S3.