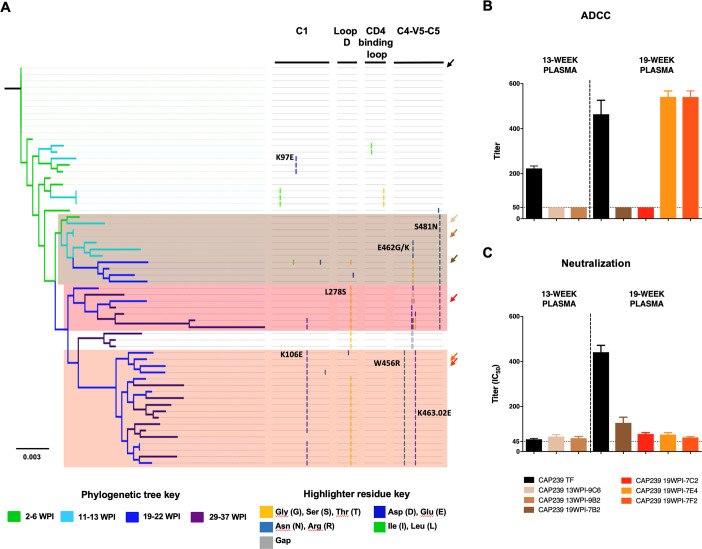
Fig 2. Impact of ADCC and neutralization responses on evolutionary pathways in CD4-binding site regions of CAP239 viruses.
(A) A maximum-likelihood tree of the 61 CAP239 full-length env single genome sequences was constructed, and rooted on the T/F virus. Branches were colour-coded according to the time-point from which the sequence was sampled. Three distinct evolutionary pathways were observed following detection of nAbs. Arrows indicated sequence used to generate infectious molecular clones containing the env of the T/F (black) as well as six representative sequences from the three pathways were constructed (identified by arrows coloured according to pathway). Two of the clones (CAP239 13WPI-9C6 and CAP239 13WPI-9B2) were representative of 13 week viruses; while four viruses (CAP239 19WPI-7B2, CAP239 19WPI-7C2, CAP239 19WPI-7F2 and CAP239 19WPI-7E4) represented 19 week viruses. (B) Env-IMCs were tested against contemporaneous plasma was tested for ADCC activity. (C) Env-IMCs were tested against contemporaneous plasma for neutralization activity. Points on ADCC and neutralisation plots indicate the mean of ≥three independent experiments.