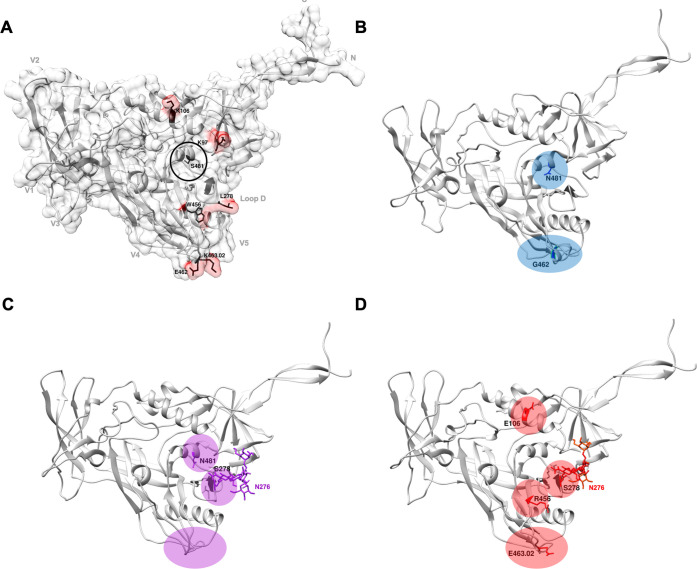
Fig 3. Models of the CAP239 gp120: effect of ADCC and neutralization escape mutations on Env structure.
Trimer models of CAP239 T/F and Envs from each of the three evolutionary pathways were constructed using Modeller [53] and the best scoring model was visualised using Chimera [54]. A single gp120 monomer from the trimer of the T/F or Envs from each pathway were used for further analysis. GlyProt [55] was used to add basic glycans (N276 in pathway two and three). (A) The contact surface was modelled onto the CAP239 T/F gp120 (grey) with any key residue surfaces shown in red. Structural landmarks (grey) and key residues in CAP239 viral evolution (black) are labelled on the CAP239 TF gp120. Changes characteristic of evolution are shown in (B) blue for pathway one (CAP239 19-7F4), (C) purple for pathway two (CAP239 19-7C2), and (D) red for pathway three (CAP239 19-7H2; pathway three).