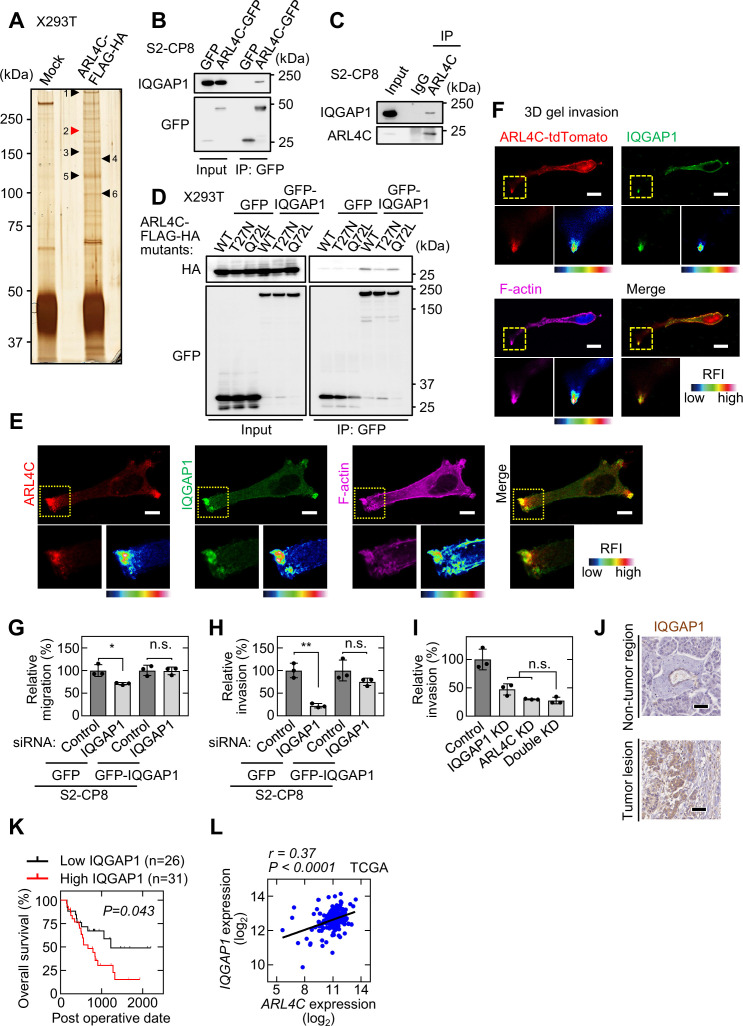
Figure 3. IQGAP1 is a novel ARL4C-interacting protein.
(A) The ARL4C-interacting proteins in X293T cells were analyzed by mass spectrometry. The results are listed in Supplementary file 1 table 2 and Source data 2. Arrowheads indicate the identified proteins, including IQGAP1 (red). (B,C) Lysates of S2-CP8 cells expressing ARL4C-GFP (B) or S2-CP8 WT cells (C) were immunoprecipitated with anti-GFP antibody (B) or anti-ARL4C antibody (C), and the immunoprecipitates were probed with the indicated antibodies. (D) Lysates of X293T cells expressing the indicated proteins were immunoprecipitated with anti-GFP antibody, and the immunoprecipitates were probed with the indicated antibodies. (E) S2-CP8 cells were stained with the indicated antibodies. Images of ARL4C and IQGAP1 were merged. (F) S2-CP8 cells expressing ARL4C-tdTomato were subjected to a 3D collagen I gel invasion assay and were stained with the indicated antibodies. Images of ARL4C and IQGAP1 were merged. (G,H) S2-CP8 cells expressing GFP or GFP-IQGAP1 were transfected with the indicated siRNAs and subjected to migration (G) and invasion (H) assays. Migratory and invasive abilities are expressed as the percentage of the same cells transfected with control siRNA. (I) S2-CP8 cells depleted of the indicated proteins were subjected to an invasion assay. Invasive activities are expressed as the percentage of control cells. (J) PDAC tissues were stained with anti-IQGAP1 antibody and hematoxylin. (K) The relationship between overall survival and IQGAP1 expression in PDAC patients was analyzed. (L) Scatter plot showing the correlation between the mRNA expression levels of ARL4C (X-axis) and IQGAP1 (Y-axis) in pancreatic cancer patients obtained from TCGA datasets using the R2: Genomics Analysis and Visualization Platform. r indicates the Pearson’s correlation coefficient. (G-I) Data are shown as the mean ± s.d. of three biological replicates. p Values were calculated using a two-tailed Student’s t-test (G,H) or one-way ANOVA followed by Bonferroni post hoc test (I). (K) The data were analyzed by Kaplan–Meier survival curves, and a log-rank test was used for statistical analysis. (E,F) The regions in the yellow dashed squares are shown enlarged in the left bottom images. The right bottom images are shown with a false color representation of fluorescence intensity. More than 50 cells were imaged and the representative image is shown. False color representations were color-coded on the spectrum. Scale bars in (E) 10 μm; (F) 20 μm; (J) 50 µm. KD, knockdown. RFI, relative fluorescence intensity. n.s., not significant. *, p < 0.05; **, p < 0.01. See Figure 3—source data 1.