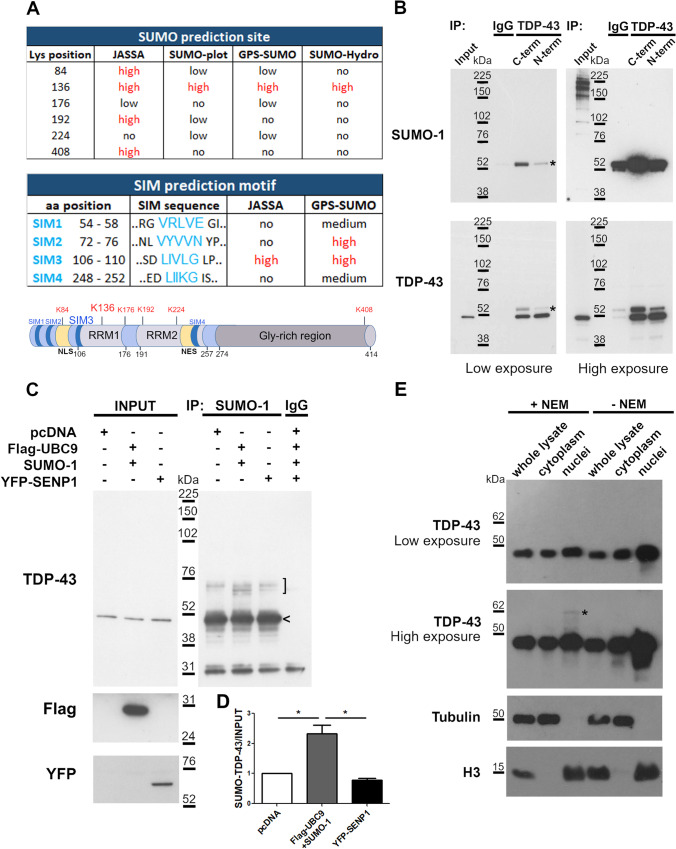
Fig. 1.
Characterization of TDP-43 protein SUMOylation. (a) In silico prediction analysis of TDP-43 SUMOylation sites (top table) and SIM motifs (bottom table) performed by different bioinformatic tools as indicated (the “high” score is indicated in red). Schematic representation of TDP-43 protein with the putative lysine (K136) and SIM motif (SIM3) predicted with a “high” score by all programs is shown. (b) Representative WB images of immunoprecipitation (IP) assay on SK-N-BE cell lysates with NEM reagent performed with two anti-TDP-43 antibodies recognizing the N-term or C-term TDP-43 domain and immunoblotted for SUMO-1 and TDP-43. IgG was used as negative control for IP (n = 3 independent experiments); asterisk, SUMOylated TDP-43 protein. (c) WB images showing IP assay on SK-N-BE cell lysate transfected with Flag-UBC9 and SUMO-1 or with YFP-SENP1 constructs. IP was performed using the anti-SUMO-1 and IgG (negative control) antibodies (n = 3 independent experiments; bracket, SUMOylated forms of TDP-43 protein; arrowhead, recovered TDP-43 protein non-covalently bounded to SUMO-1. (d) Image quantification of WB data presented in (c) (mean ± s.d; One-way ANOVA and Tukey post hoc test; n = 3; *p < 0.05). (e) Representative WB images of nucleo-cytoplasm fractionation of SK-N-BE cell lysates with or without NEM (n = 5 independent experiments; asterisk, the SUMOylated TDP-43 protein)