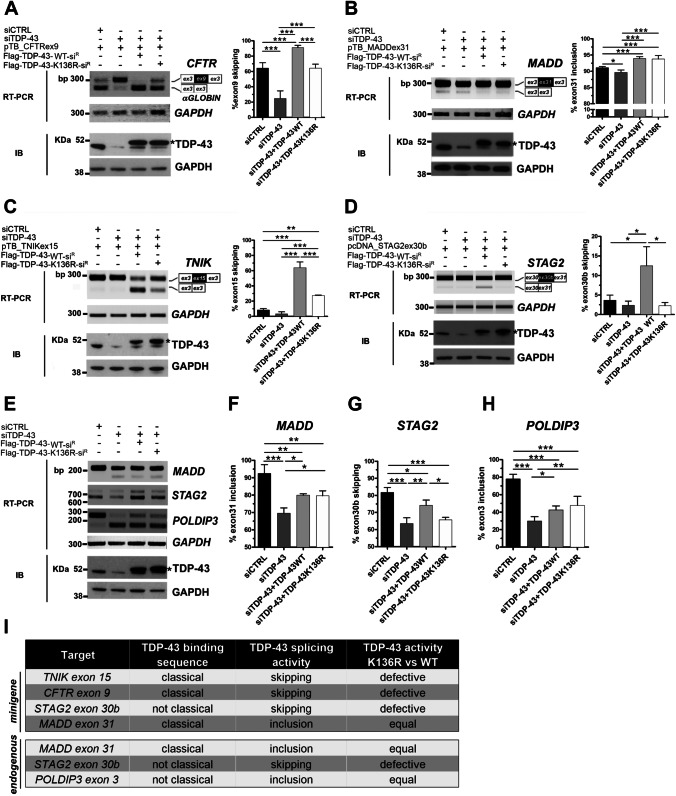
Fig. 3.
Splicing activity of the SUMO-resistant TDP-43 K136R protein. (a–d) Representative RT-PCR (Upper panels) and immunoblot (IB) (Lower panels) images of minigene splicing assays in HEK293T cells knocked-down for TARDBP gene and co-transfected with the siRNA resistant Flag-TDP-43 WT or K136R constructs and the pTB_minigenes CFTRex9 (a), MADDex31 (b), TNIKex15 (c) or the pcDNA_minigene STAG2 (d) as indicated. GAPDH was used for data normalization in both RT-PCR and IB assays. Asterisk, exogenous Flag-tagged TDP-43 WT or K136R proteins. Densitometric analyses of CFTRex9 skipping (a), MADDex31 inclusion (b), TNIKex15 skipping (c) and STAG2ex30b skipping (d) data from the minigene assays (mean ± s.d.; One-way ANOVA and Tukey post hoc test; n = at least 3 independent experiments; *p < 0.05; **p < 0.01; ***p < 0.001). (e) Representative RT-PCR (Upper panels) and IB images (Lower panels) of endogenous MADD, STAG2 and POLDIP3 alternative splicing in HEK293T cells, knocked-down for TARDBP and transfected with the siRNA-resistant Flag-TDP-43 WT or K136R constructs. GAPDH was used for sample normalization. (f–h) Densitometric analyses of endogenous MADDex31, STAG2ex30b and POLDIP3ex3 splicing (mean ± s.d.; One-way ANOVA and Tukey post hoc test; n = at least 3 independent experiments; *p < 0.05; **p < 0.01; ***p < 0.001). (i) Summary table of the splicing activity of TDP-43 K136R versus the WT protein on minigenes and endogenous gene targets. The types of consensus binding sequence and splicing event for the analysed target are also reported for comparison