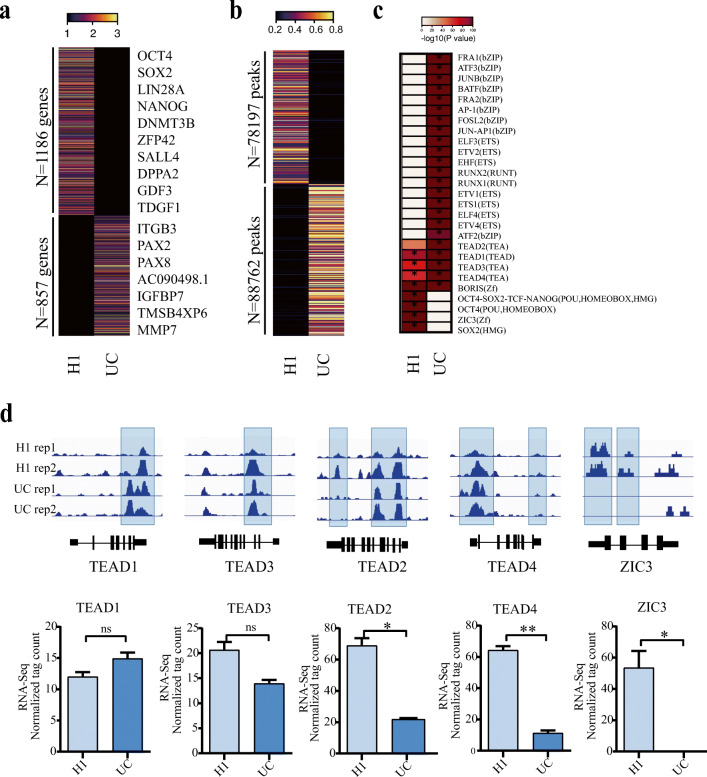
Fig. 1.
Screen candidate reprogramming factors by analysing chromatin accessibility and transcriptome data. a Heatmap of RNA-seq analysis results of UCs and hESCs (H1). There are 1186 highly expressed genes in hESCs (H1) and 857 highly expressed genes in UCs. b ATAC-seq chromatin status. In order to make the data clearer, the sequencing results of the two replicates of UCs and hESCs (H1) were merged into one dataset using the mean value. There are 88,762 peaks opened in UCs but closed in hESCs (H1) and 78,197 peaks closed in UCs but opened in hESCs (H1). c DNA motif enrichment analysis of two groups of ATAC-seq data by HOMER [27]. d Chromatin landscape and gene expression value of the target genes (UC rep1 and UC rep2 represent ATAC-seq data from two donors). RNA-seq data are presented as mean ± SEM, n = 2. ns P>0.05, *P < 0.05, **P < 0.01, unpaired two tailed student-t-test