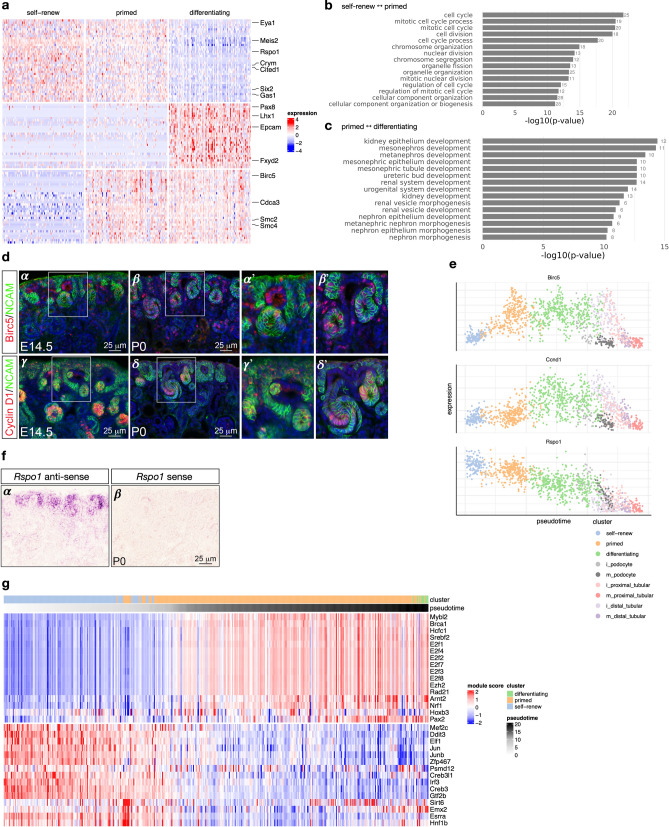
Figure 3.
Transcriptional signatures of self-renewing, primed and differentiating nephron progenitor cells. (a) shows differentially expressed genes on a heatmap of 100 random cells for each of the “self-renew”, “primed” and differentiating clusters, with key genes annotated on the right. (b) and (c) show the 20 most-enriched Gene Ontology terms for genes differentially expressed between self-renewing and primed NP cells, and between primed NP cells and differentiating cells, respectively. (d) Immunofluorescence on kidney sections from embryonic day 14.5 (E14.5) and postnatal day 0 (P0) mice using anti-BIRC5 (α - β′) and anti-Cyclin D1 (γ - δ’) antibodies (red). Nephron progenitors and their early epithelial derivatives were detected using an antibody against anti-Neural cell adhesion molecule (NCAM; green). Nuclei were counterstained with DAPI (blue). Scale bar, 25 μm. The sub-panels α′, β′, γ′ and δ′ are close-ups of the areas indicated by the white boxes. (e) Expression of Birc5, Ccnd1 and Rspo1 across pseudotime; colors indicate cell clusters. (f) In situ hybridization on cryosections of P0 kidneys confirms the expression of Rspo1 in nephron progenitors and their early epithelial derivatives (α). No signal was detected with sense probe hybridization (β). Images are representative of three independent experiments. Scale bar 25 μm. (g) Inferred regulatory module activity based on SCENIC50 across pseudotime for predominantly self-renewing and primed nephron progenitor cells.