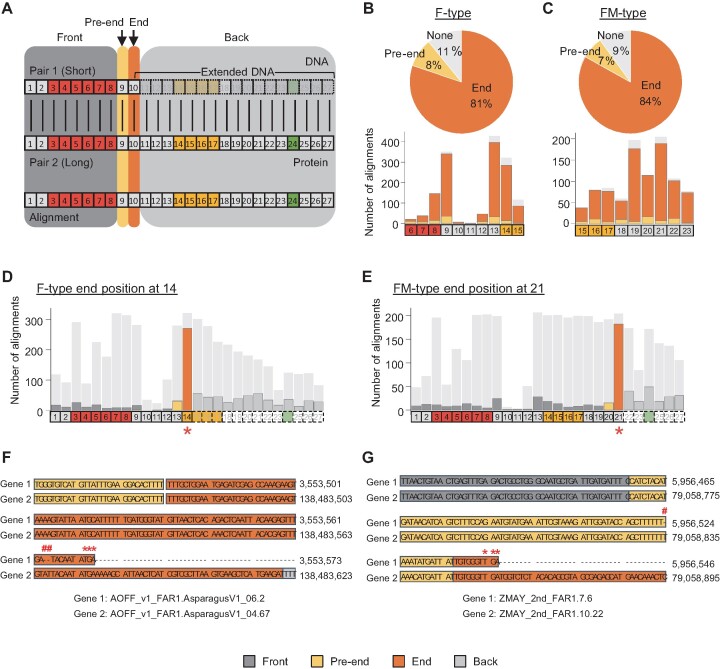
Figure 3.
Distribution of accumulated frameshift or nonsense mutations in FAR1 (F-type) and FAR1-MULE (FM-type) pairs. (A) Motif positions of pairs and alignment result of the pair divided into four sections. Shorter and longer gene structures were designated Pair 1 and Pair 2, respectively. Motif containing the stop codon of Pair 1 was termed the ‘end’ motif site, and the motif directly upstream of ‘end’ was termed ‘pre-end’. Other motifs upstream of the ‘pre-end’ motif site were called ‘front’ motif sites, while motifs downstream of the ‘end’ motif site were called ‘back’ motif sites. Red, yellow and green boxes represent motifs of FAR1, MULE and SWIM domains, respectively. (Band C) The number of mutations in pre-end (yellow) or end (orange) motif sites for each set of alignments (light gray) with end motif sites at 6–15 for F-type and 15–23 for FM-type is illustrated as bar plots. The proportion of genes with mutation occurrences in the pre-end or end positions are shown as a pie chart. ‘None’ refers to alignments with no mutations in pre-end or end motif sites. (Dand E) Specific examples show the mutation distribution in F- and FM-type alignments with end positions at 14 and 21, respectively. Light gray bars with no bar lines show the total number of motifs in each position of alignments while the colored bars indicate the number of alignments with mutations for each position. (B–E) The x-axis is the positional information of motifs. Red, yellow and green boxes represent motif positions of FAR1, MULE and SWIM domains, respectively. Solid boxes denote motif positions contained in the shorter pair, while dotted boxes are not part of the shorter pair but included in the alignments. Colors of bars delineate the motif position in relation to the end motif (asterisk) of the shorter pair. (Fand G) Duplication pairs with a frameshift mutation within the end motif site or pre-end motif site causing premature termination are shown. Top genes are the shorter of the paired genes. Hashtag (#) represents the site where frameshift mutation is predicted and asterisks show the stop codon where premature termination occurred. Colors indicate the motif position in relation to the end motif site (orange) of the shorter pair.