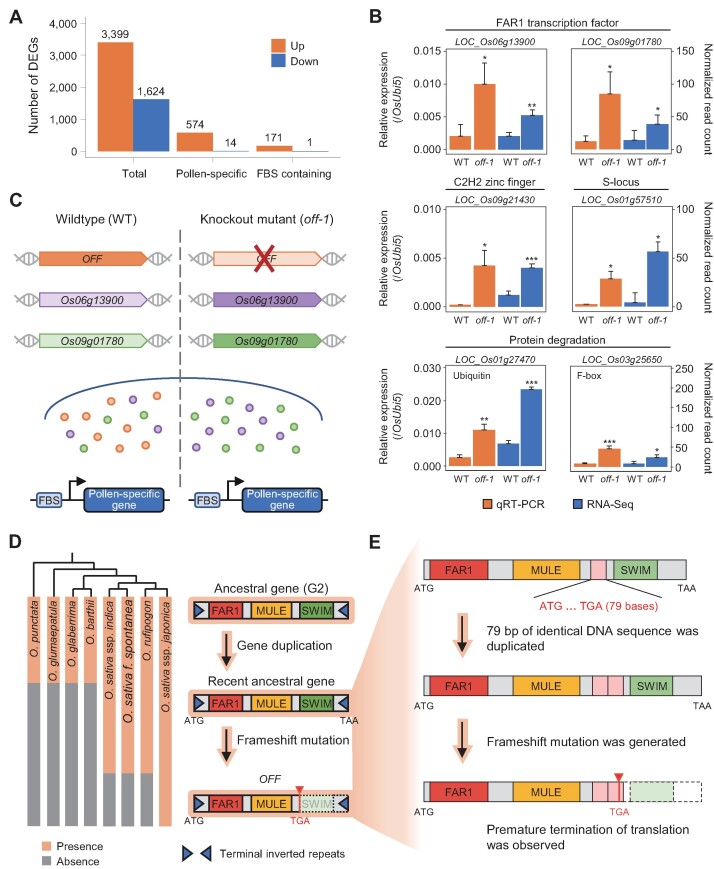
Figure 5.
Transcriptional regulation of rice pollen by the mutation of Oryza sativa FAR1 related to Fertility (OFF) gene and its evolutionary history. (A) The number of differentially expressed genes (DEGs) is shown as a bar graph. FBS indicates FAR1 binding site (CACGCGC). Colors of bars show upregulated (orange) and down-regulated (blue) genes. (B) qRT-PCR validation of up-regulated FAR1 transcription factors and four regulatory genes. Relative expression values (orange) and normalized RNA-seq read counts (blue) are shown. The rice ubiquitin 5 gene (OsUbi5, LOC_Os01g22490) was used to calculate the relative expression value as an internal control. The error bars indicate standard deviation. * means P < 0.05; ** P < 0.01; *** P < 0.001. (C) Hypothetical model of OFF function in rice pollen. Colored dots indicate translated OFF (orange), Os06g13900 (purple) and Os09g01780 (green). Opaque colors represent high expression while lighter colors indicate low expression of genes. (D) The global evolutionary mechanism is illustrated. The recent parent gene derived from an ancestral FAR1 gene, common to all Oryza species in Group 2, was duplicated in O. sativa. Orange stripes on the left highlight Oryza subspecies that share evidence of the target gene shown on the right, while gray stripes indicate absence of such gene evidence. (E) The specific evolutionary process of OFF is depicted. After duplication of the recent parent gene, a 79-bp DNA sequence was duplicated, generating a frameshift mutation. Premature termination of protein translation (red arrowhead) was observed, and a function was newly acquired.