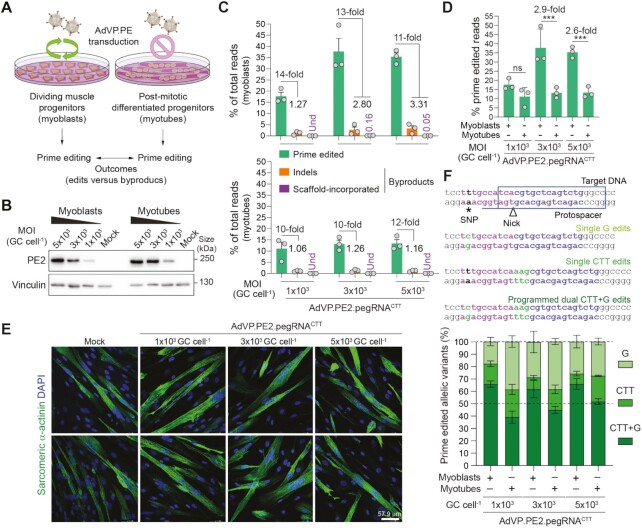
Figure 4.
Assessing the impact of cellular replication on prime editing performance. (A) Experimental set-up for characterizing prime editing in dividing versus post-mitotic cells. (B) Analyses of PE2 expression in muscle cells. Muscle progenitor cells were transduced before and after differentiation with AdVP.PE2.pegRNACTT at the specified MOI and, 2 days later, PE2 expression levels were assessed by western blotting. Cas9- and vinculin-specific antibodies detected target (PE2) and loading control proteins, respectively. (C) Characterization of prime editing in dividing versus post-mitotic cells. The frequencies of prime edited and byproduct alleles in myoblasts and myotubes (top and bottom graphs, respectively) exposed to different MOI of AdVP.PE2.pegRNACTT, were determined through CRISPResso2 analysis at 3 days post-transduction. Byproducts consist of small deletions and insertions (indels; orange bars) and pegRNA scaffold-derived insertions (violet bars). Graph presents mean ± s.e.m. of three biological replicates; Und, undetected. (D) Prime editing frequencies in mitotic versus post-mitotic cells. Aggregated prime editing frequencies indicated in panel C highlighting differences in PE2:pgRNACTT activity in myoblasts versus myotubes. Bars and error bars correspond to mean and s.d, respectively. Significance between datasets was calculated with two-way ANOVA followed by Sidak's test for multiple comparisons; ***0.0001 < P < 0.001; P > 0.05 was considered non-significant (ns). (E) Differentiation of AdVP.PE2.pegRNACTT-treated myoblasts. The differentiation capacity of vector-transduced myoblasts was ascertained by immunofluorescence microscopy analysis for the late muscle-specific marker sarcomeric α-actinin after incubating the cells in low-mitogen medium. Nuclei in syncytial myotubes were identified by DAPI staining. Mock-transduced myoblasts served as controls. Two representative micrographs for each experimental condition are shown. (F) Parsing of prime-edited allele variants resulting from a composite prime editing design. Myoblasts and RPE-Fucci cells are homozygous for a SNP in the region complementary to the pegRNACTT RT template permitting assessing composite (CTT + G) versus single (CTT or G) edits instructed by PE2:pegRNACTT complexes. Genomic sequences before and after the delivery of PE2:pegRNACTT complexes into cells containing a SNP in the region complementary to the RT template (magenta nucleotides), are depicted (top panel). Prime editing outcomes instructed by pegRNACTT correspond to alleles containing A > G substitutions, CTT insertions or both modifications. Replicating myoblasts and post-mitotic myotubes were transduced with the all-in-one vector AdVP.PE2.pegRNACTT. Discrimination and quantification of the different target alleles was performed via next-generation deep sequencing analysis on genomic DNA isolated at 3 days post-transduction (bottom panel). Data are plotted as mean ± s.e.m. of three biological replicates.