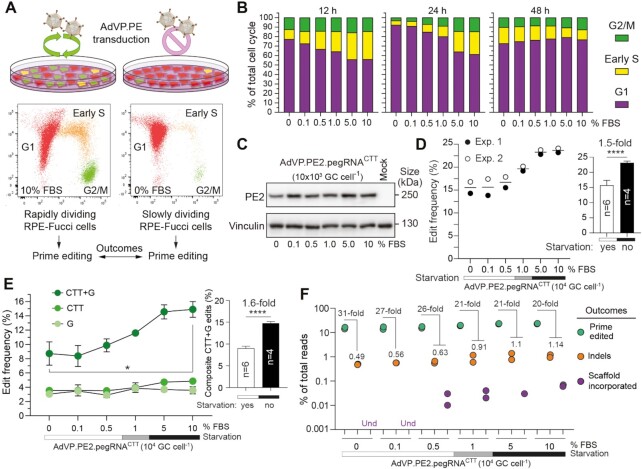
Figure 5.
Characterization of prime editing in rapidly versus slowly dividing cell populations. (A) Experimental set-up for characterizing prime editing in rapidly versus slowly dividing cell populations. RPE-Fucci cell cultures containing varying proportions of cycling and non-cycling cells were established by applying a serum gradient. Fucci reporters in these cells trace cell fractions in G1, early S or G2/M phases, permitting the monitoring of cell division-to-prime editing rates upon AdVP.PE2.pegRNACTT transduction. (B) Tracking cell cycling and prime editing activities in RPE-Fucci cell cultures. Cell cycle analysis was done at the indicated timepoints on RPE-1 cells exposed to various FBS concentrations by flow cytometry. Parallel cultures of RPE-Fucci cells were transduced with AdVP.PE2.pegRNACTT at 12 hours after FBS treatments initiation. (C) Analysis of PE2 expression in RPE-1 cells. PE2 expression levels were determined by western blotting using Cas9- and vinculin-specific antibodies for detecting PE2 and loading control proteins, respectively. (D) Quantification of prime editing outcomes in RPE-1 cells. Frequencies of prime editing and prime editing collateral events were measured by CRISPresso2 analysis at 60 hours post-transduction. Datapoints derive from two independent experiments carried out throughout different days. Bars represent mean ± s.d. of prime editing activities in RPE-Fucci cells exposed to serum starvation (0, 0.1, and 0.5% FBS) or regular culture conditions (5 and 10% FBS). Unpaired two-tailed Student's t test ****P < 0.0001. (E) Parsing of prime-edited allele variants resulting from the composite prime editing design. Statistical significance was assesed by Student's t tests; *0.01 < P < 0.05; ****P < 0.0001. (F) Relationship between prime edited alleles and byproduct variants in AdVP-transduced RPE-Fucci cells. Und, undetected.