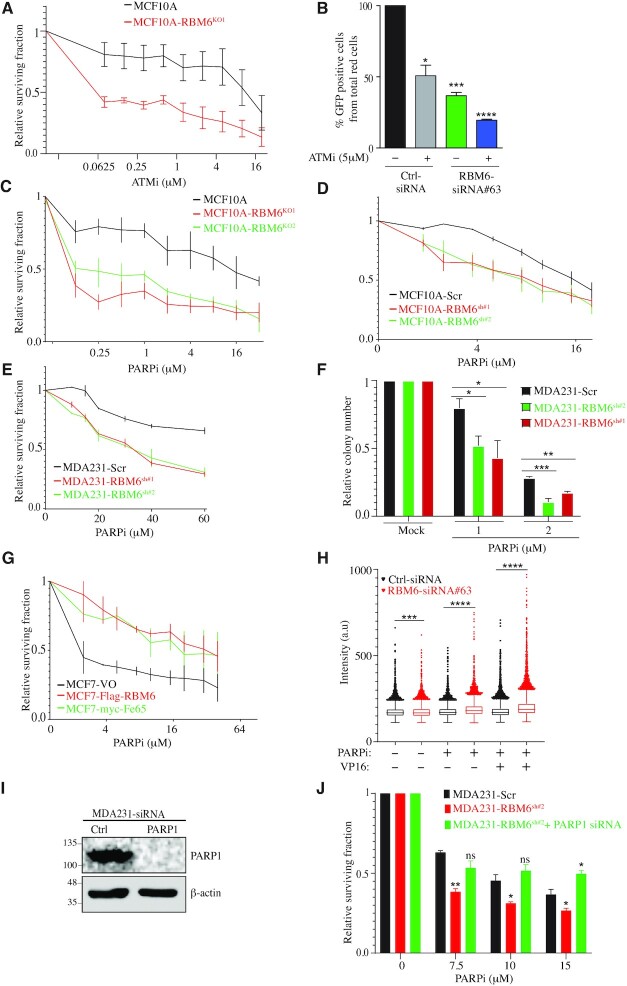
Figure 7.
RBM6 depletion sensitizes cells to ATM and PARP inhibition. (A) MCF10A RBM6-KO1 cells are hypersensitive to ATM inhibition. Cell viability was determined by the CellTiter 96® proliferation assay. (B) ATM inhibition exacerbate the HR deficiency in RBM6 depleted cells. U2OS-HR cells were transfected with siRNA against RBM6 and left untreated or treated with 5μM ATMi and tested for HR efficiency as described in Figure 2A. (C, D) PARP inhibition sensitizes MCF10A RBM6-KO1 and KO2 (C) or RBM6-shRNA#1 and #2 (D) cells. Cell viability was assessed by the CellTiter 96® proliferation assay. (E) As in (D), except that MDA231 cells were used. (F) MDA231 cells constitutively depleted of RBM6 were treated with PARP inhibitor. Cell viability was measured by clonogenic assay. (G) MCF7 cells expressing Flag-RBM6 or myc-Fe65 were treated with PARP inhibitor and cell viability was measured by CellTiter 96® proliferation assay. Data presented are mean of three independent experiments ± SD. (H) Image-based quantification of PARPi-induced PARP1 trapping. MDA231 cells were treated with 10 μM olaparib and 40 μM VP16 for 4 h as indicated, pre-extracted on ice in 0.2% Triton X-100 for 2 min to remove soluble, non-chromatin-bound proteins, and stained for PARP1 and DNA content. Experiment was performed in triplicates. (I) Western blot analysis shows knockdown of PARP1 in MDA231 cells. β-Actin was used as a loading marker. (J) PARP1 was depleted in RBM6-deficient MDA231 cells and PARP inhibition effect was measured by CellTiter 96® proliferation assay. Data presented are mean of three independent experiments ± SD. *P< 0.01, **P < 0.001, ***P < 0.0001, ****P < 0.00001. All data presented are mean of three independent experiments ± SD.