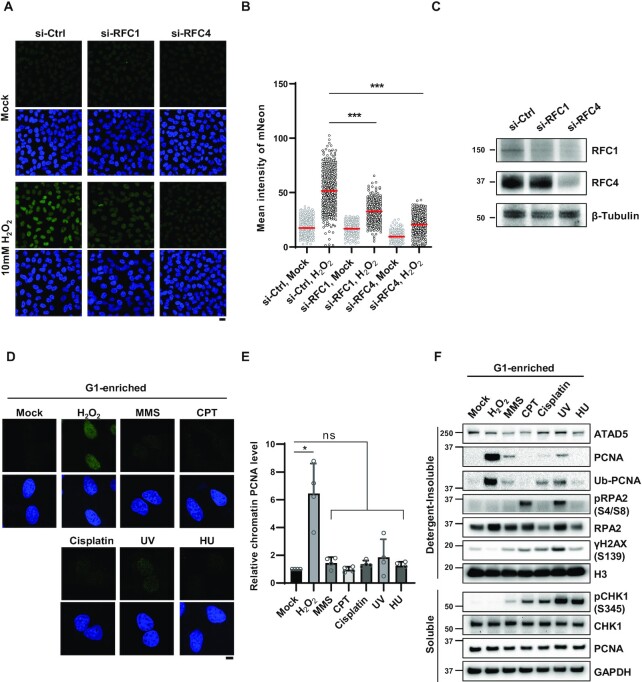
Figure 2.
ATAD5 accumulation upon oxidative DNA damage depends on RFC1. (A–C) HeLa-ATAD5mNeonGreen-AID cells transfected with small interfering RNAs (siRNAs) as indicated for 48 h. Cells were treated with 10 mM H2O2 for 10 min, detergent-pre-extracted and then fixed for detection of mNeonGreen signal. (A) Representative images are shown; scale bar: 20 μm. (B) Quantification of mNeonGreen signals. Three independent experiments were performed and one representative result is displayed. Red bar indicates mean value. (C) Results of immunoblotting of whole cell extracts prepared 48 h after transfection. (D and E) HeLa cells were treated with nocodazole for 5 h, shaked-off and released into fresh medium followed by incubation for 4 h before treatment with DNA-damaging agents under conditions used in Figure 1D and E. After damage treatment, cells were, pre-extracted with detergent before fixation and immunostained with an anti-PCNA antibody. (D) Representative images are shown; scale bar: 10 μm. (E) The intensity of PCNA signal was quantified. PCNA intensity of four independent experiments are normalized. Error bars represent standard deviation of the mean (n = 4). (F) U2OS-ATAD5AID cells previously reported (14) were enriched at G1 phase by treatment with PD 0332991 and PHA-767491, and treated with DNA-damaging agents under conditions used in Figure 1D and E. After drug treatment, detergent-soluble and -insoluble proteins were fractionated and subjected to immunoblotting. (B and E) Statistical analysis: two-tailed unpaired (B) and paired (E) Student's t-test; ***P < 0.005, *P < 0.05, and ns: not significant.