Figure 1.
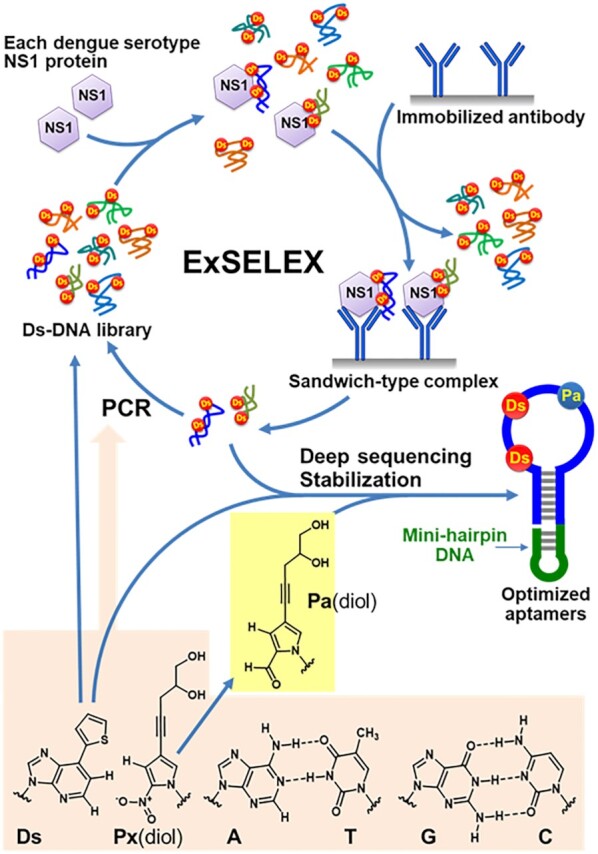
ExSELEX scheme to generate UB-DNA aptamers targeting each DEN-NS1 serotype or sub-serotype variant using an aptamer−antibody sandwich method. The Ds-containing DNA library is mixed with each target DEN-NS1. The DNA–protein complexes are captured with the immobilized anti-NS1 antibody. After washing to remove the unbound DNA species, the bound DNA species are recovered and subjected to PCR amplification involving the Ds–Px pair as a third base pair, to obtain the enriched DNA library for the next round of selection. After several ExSELEX rounds, the sequences in the enriched DNA libraries are determined by deep sequencing, and the aptamer candidates are optimized and stabilized by adding an extraordinarily stable mini-hairpin DNA. One of the aptamer candidates contained the Px base, which resulted from the mutation from the natural base to Px during PCR in ExSELEX. Since the Px nucleoside is unstable under the DNA chemical synthesis conditions, the Px(diol) nucleosides in the aptamers were replaced with the Pa(diol) nucleotide for the synthesis. The Ds–Px pair is a hydrophobic unnatural base pair without any clear hydrogen-bonding interactions between the pairing bases (59).
