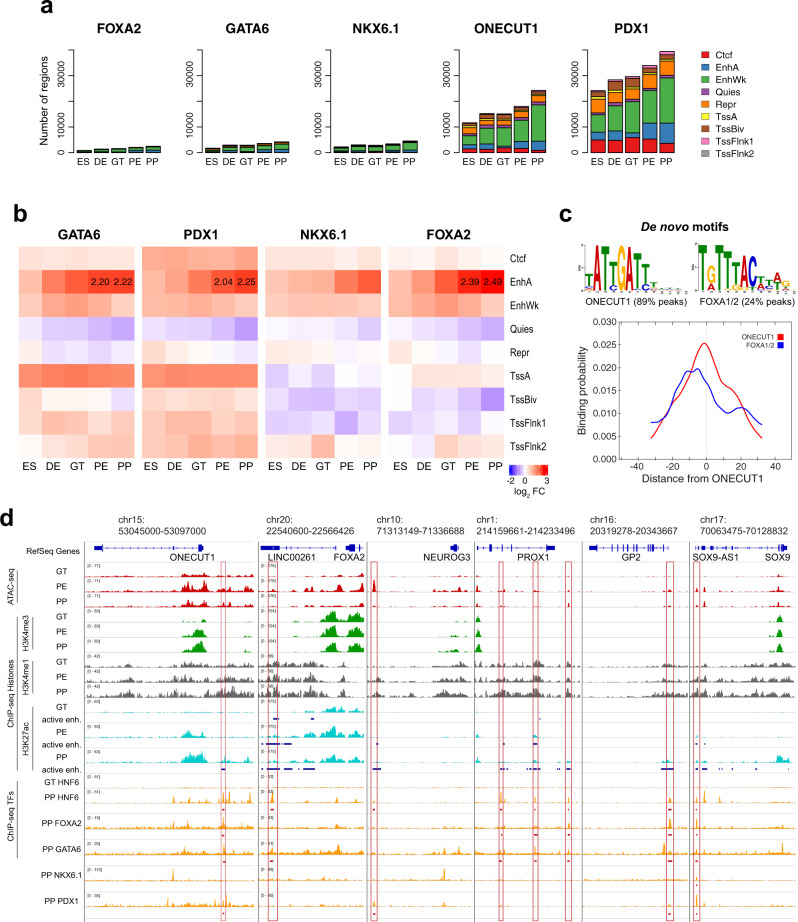
Fig. 4. ONECUT1 cobinding during early pancreas differentiation regulation.
a Overlap of peaks of distinct TFs on chromatin states as delineated by chromHMM. These states include active enhancers (EnhA), weak/poised enhancers (EnhWk.), repressed regions (Repr.), active promoters (TssA), poised promoters (TssBiv) and regions flanking active promoters (TssFlnk1-2). b Log2 fold change (FC) of the frequency of ONECUT1 co-binding events (ONECUT1 with either GATA6, PDX1, NKX6.1, and FOXA2) vs. ONECUT1 exclusive binding sites for distinct chromatin states and differentiation stages. Red values indicate chromatin stages over-represented in co-binding events. We highlight values with abs(log2(FC)) > 2. c We show MEME de novo motifs around ONECUT1 ChIP-seq peaks, which are found in 89% of peaks, and FOXA2 motif, which is found in 24% of peaks. Both motifs have a preference relative to the peak center, i.e., ONECUT1 is mostly localized in the peak middle, while FOXA2 is located 9–10 bps downstream of peak center. d IGV Genome browser view of ONECUT1 and PE/PP expressed genes with ChIP-seq and ATAC-seq profiles from GT, PE, and PP stages. We highlight ONECUT1 peaks overlapping with active enhancers (blue tracks below H3K27ac tracks) gained at PE or PP stage, as well as co-binding partners.