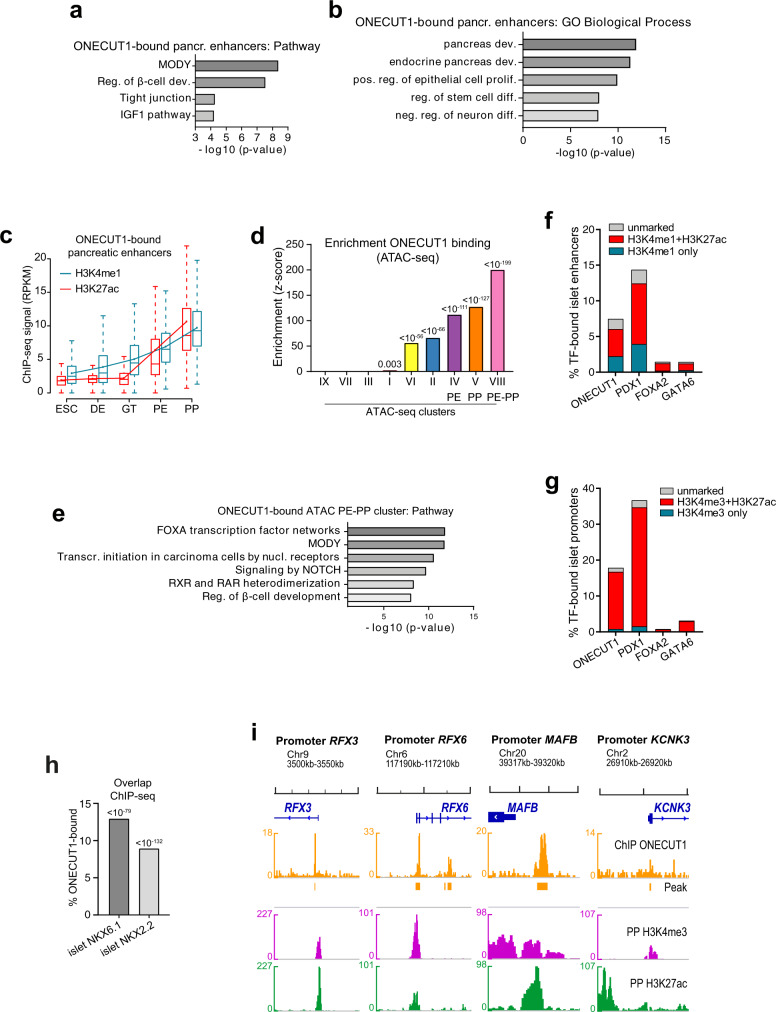
Fig. 5. Pancreatic and islet enhancers are bound by ONECUT1.
a, b Enrichment analysis (GREAT36) of ONECUT1-bound pancreatic enhancers (FG/PE and PE clusters from ref. 29) for “pathway terms” (a) from the MSigDB pathway genes subset CP and for “GO Biological Process” (b) from the Gene Ontology Consortium (Binomial test; one side). c Distribution of stage-specific H3K4me1 and H3K27ac ChIP-seq signal at ONECUT1-bound pancreatic enhancers (FG/PE and PE clusters from ref. 29). Boxplots depict median (center), interquartile range (box) and extreme values (whiskers), point and connecting line mean value. d Binding enrichment (z-score) test of ONECUT1 (ChIP-seq, PP stage) in distinct OC clusters of HUES8 ATAC-seq (z-test; one side). e Enrichment analysis (GREAT36) of ONECUT1-bound genes overlapping with PE/PP ATAC-seq (cluster VIII) for “pathway terms” (Binomial test; one side). f, g Overlap between ChIP-seq peaks of ONECUT1 (PP stage) and depicted TFs with islet enhancers (f) and islet promoters (g) together with distinct histone modification as defined by ref. 45. h Overlap between ONECUT1 ChIP-seq peaks (PP stage) with islet TF peaks (NKX6.1 and NKX2.2 ChIP-seq from human islets45; intersection test, one side84). i Interactive Genome Viewer (IGV) plots illustrates ONECUT1 binding peaks and histone marks in promoter regions for selected endocrine genes. Genomic locations refer to human genome assembly GRCh37 (Hg19).