Figure 6.
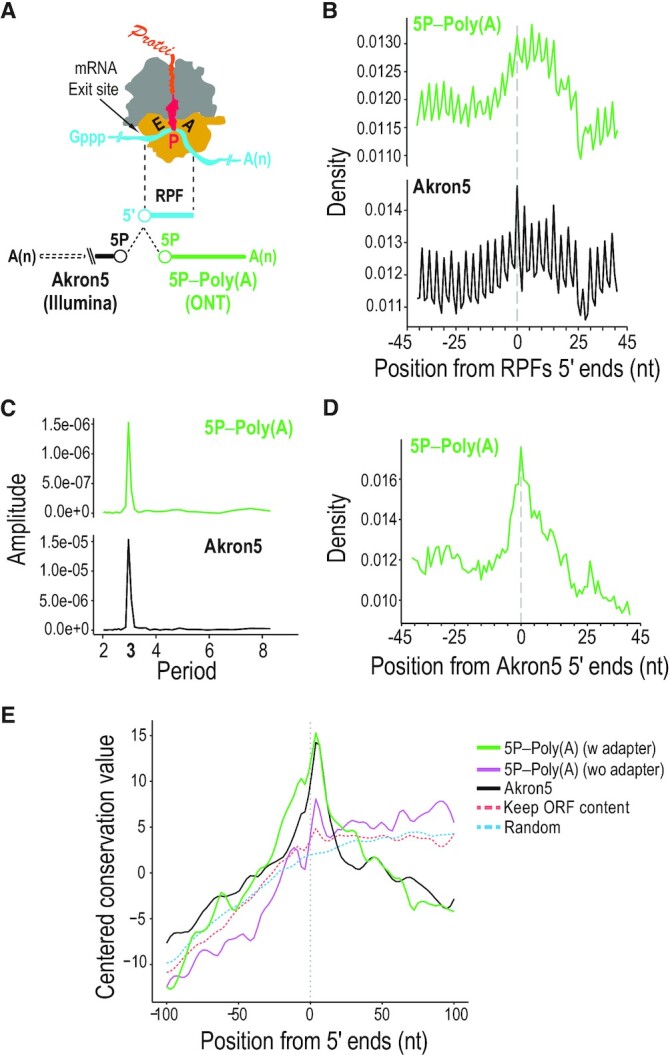
Cotranslational mRNA decay identified with TERA-Seq. (A) Schematic of elongating ribosome with ribosome-protected fragment (RPF) and relative position analyses of 5′ ends from TERA-Seq (5P–Poly(A); ONT) and Akron-Seq (Akron5; Illumina) to the 5′ ends of RPFs. E, tRNA-exit site; P, peptidyl-tRNA site; A, aminoacyl-tRNA-binding site; yellow, 40S subunit with mRNA channel; grey, 60S subunit with polypeptide channel; red, peptidyl-tRNA attached to nascent protein; Gppp, 5′ cap; A(n), poly(A) tail; 5P, 5′ monophosphate. (B) Density plots of 5′ ends distances from indicated libraries relative to RPF 5′ ends (centered at position 0) in coding regions. RPF, ribosome-protected fragment. nt, nucleotide. (C) Discrete Fourier transformation of read density around RPFs for indicated libraries. (D) Density plot of 5′ ends distances from TERA-Seq (5P–Poly(A)) relative to Akron-Seq (Akron5). (E) Evolutionary conservation for 100 vertebrates (PhastCons) upstream and downstream of 5′ ends of mRNAs identified from the 5P-Poly(A) TERA-Seq library; adapted (green) and non-adapted (purple) reads and reads from Akron5 Illumina library (black) are shown. A random control maintaining the nucleotide and open reading frame (ORF) context of 5P-Poly(A) 5′ ends (dashed orange) and a completely random control (dashed blue) are shown.
