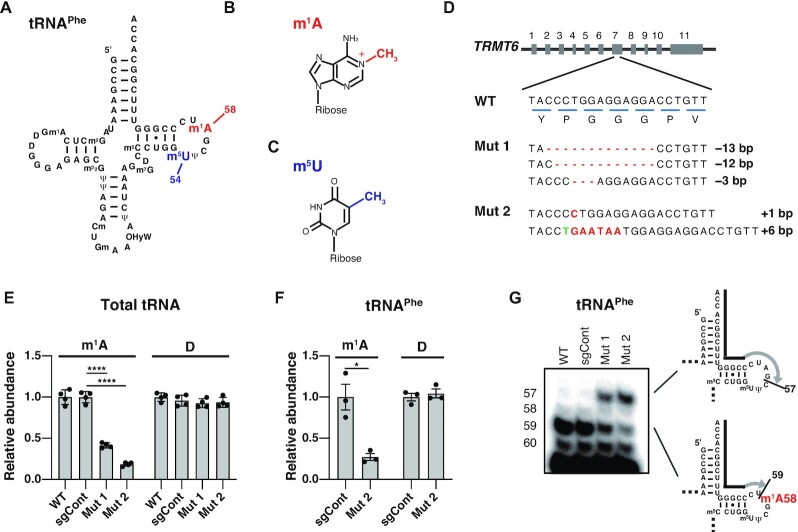
Figure 1.
Construction of TRMT6 mutant cell lines. (A) Secondary structure of the human cytoplasmic tRNAPhe with modified nucleosides: N1-methyladenosine (m1A), 5-methyluridine (m5U), N2-methylguanosine (m2G), dihydrouridine (D), N2,N2-dimethylguanosine (m22G), pseudouridine (Ψ), 2′-O-methylcytidine (Cm), 2′-O-methylguanosine (Gm), hydroxywybutosine (OHyW), 7-methylguanosine (m7G), and 5-methylcytidine (m5C). (B) Chemical structure of m1A. (C) Chemical structure of m5U. (D) TRMT6 alleles in TRMT6 mutant cells. Red lines indicate deletions. Red letters indicate insertions. Green letter indicates base exchange. Detection of three TRMT6 alleles in TRMT6 mutant #1 may be because HEK293FT cell is a near-triploid cell. (E) LC–MS analysis of total tRNA nucleosides from cells. Dihydrouridine level is shown as a control. WT indicates wild-type HEK293FT cells. sgCont indicates a control HEK293FT cell line that was generated using negative control sgRNA that does not target the human genome. Mut 1 and Mut 2 are TRMT6 mutant cell lines illustrated in Figure 1D. Relative abundance indicates LC-MS peak area relative to WT measurements. Means ± standard error of means (s.e.m.) from n = 4 biological replicates. (F) LC–MS analysis of nucleosides made by digestion of purified tRNAPhe. Dihydrouridine level is shown as a control. Relative abundance indicates LC-MS peak area relative to sgCont measurements. Means ± s.e.m. from n = 3 biological replicates. (G) Detection of hypomodified m1A58 using primer extension of HEK293FT cell line RNA samples. The primers are shown as black lines, and nascent cDNAs synthesized from the primers are depicted as gray lines. For reverse transcription, in addition to dATP, dTTP and dGTP, dideoxyCTP (ddCTP) was used to terminate reverse transcription at tRNAPhe G57. ****P < 0.0001, *P < 0.05 by Welch's t-test.