FIGURE 1.
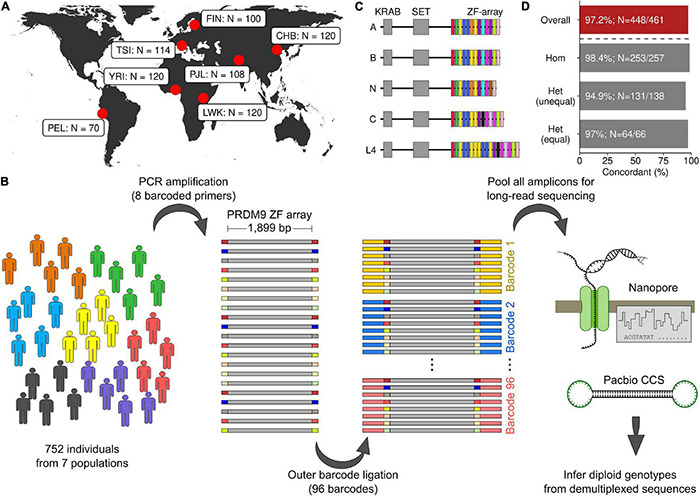
PRDM9 diploid genotyping with long-read sequencing. (A) Geographic location of seven populations in this study (YRI, Yoruba in Ibadan, Nigeria; LWK, Luhya in Webuye, Kenya; TSI, Toscani in Italia; FIN, Finnish in Finland; PEL, Peruvian in Lima, Peru; CHB, Han Chinese in Beijing, China; PJL, Punjabi in Lahore, Pakistan). (B) Schematic of amplification, barcoding, and sequencing strategy (see section “Materials and Methods”). (C) The protein domain structure of human PRDM9. The zinc finger (ZF) array of PRDM9 is a repeating array of 84-bp-long ZFs. ZF variants are indicated by different colors. Five annotated PRDM9 alleles are shown. (D) Genotyping PRDM9 gives analogous results using either Oxford Nanopore or Pacific Biosciences Circular Consensus Sequencing (CCS) (PacBio CCS). The percentage of concordant genotypes is shown. The least agreement (94.9%) is seen for individuals that are heterozygous for alleles of different lengths [Het (unequal)]. The overall concordance across all 461 individuals is 97.2%.
