FIGURE 4.
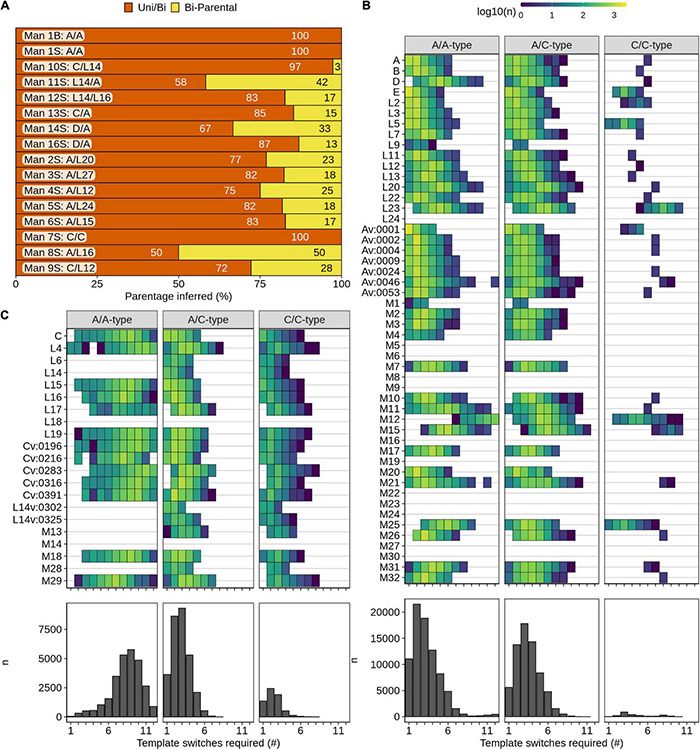
Isolated clusters of A-type and C-type PRDM9 alleles. The PRDM9 ZF array is hypervariable, and variation arises via a poorly understood mechanism. We designed and utilized an algorithm to predict the formation of any PRDM9 allele from any other based on template switching. (A) Most PRDM9 variants in human blood or sperm can be explained by template switches between the two parental PRDM9 alleles. If several combinations of parental alleles are possible, we identified the “most-likely” recombinant, which required the minimal number of template switches. For each individual, we quantified the alleles where this “most-likely” recombinant is derived from either both parental alleles (bi-parental) or where a uni- and bi-parental origin are equally possible (Uni/Bi). (B) A-type PRDM9 alleles rarely arise from C-type alleles, and (C) C-type PRDM9 alleles can arise from A-type alleles but mostly require highly complex template switches. (B,C) We searched for potential parental alleles for each A-type (B) and C-type (C) PRDM9 allele. All alleles found in human populations or in blood/sperm only were considered. Heatmaps show the number of potential parental combinations for each number of template switches. Columns represent events in A-type homozygotes, A-type/C-type heterozygotes, or C-type homozygotes. Quantitation of all events is shown in bar plots underneath.
