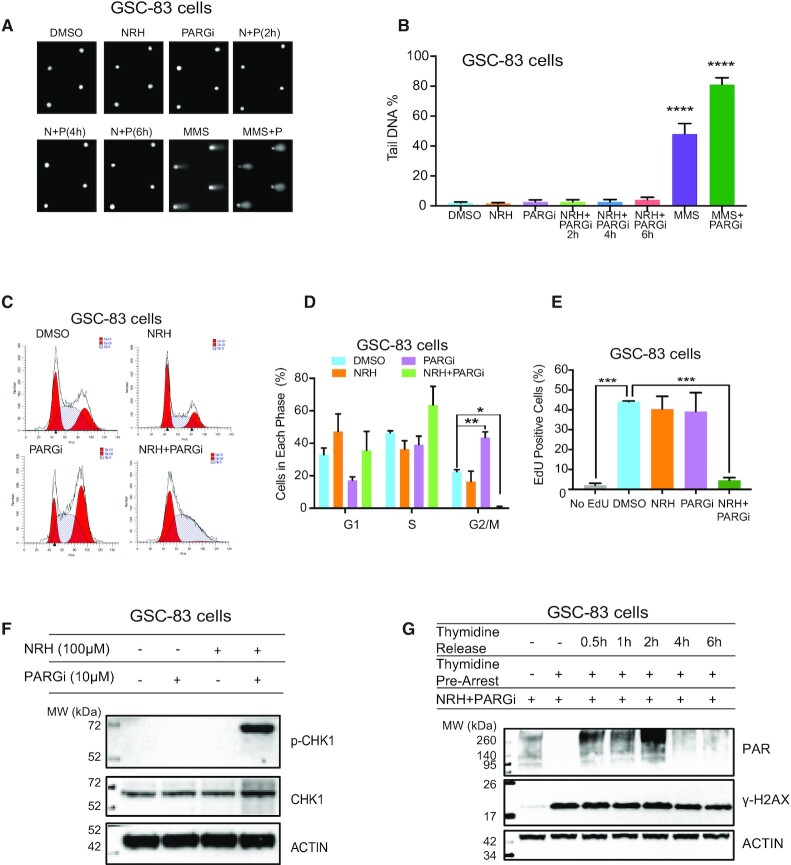
Figure 4.
PARGi-induced S-phase arrest and checkpoint activation requires enhanced cellular NAD+ from NRH exposure. (A and B) Representative Comet images (A) and quantification of genomic DNA damage (B) resulting from cellular treatment with DMSO, NRH (100 μM), PARGi (10 μM) or NRH+PARGi (N+P) for the duration indicated. GSC-83 cells were also exposed to MMS (1 mM) or MMS+PARGi (MMS+P) for 30 min. DNA damage was evaluated by the CometChip assay, reported as % Tail DNA; ****P < 0.0001 as compared to the control, DMSO. (C) Flow cytometry cell cycle analysis of GSC-83 cells 24 h after treatment with DMSO, NRH (100 μM), PARGi (10 μM) or NRH+PARGi. (D) Quantification of the cell cycle analysis of GSC-83 cells 24 h after treatment with DMSO, NRH (100 μM), PARGi (10 μM) or NRH+PARGi (*P < 0.05, **P < 0.01), showing the percentage of cells in each cell cycle phase (three separate experiments). (E) Percentage of EdU positive cells (GSC-83 cells) 24 h after treatment with DMSO, NRH (100 μM), PARGi (10 μM) or NRH+PARGi followed by 2 h EdU incorporation (***P < 0.001). (F) Immunoblot analysis of CHK1 and phosphorylated CHK1 (p-CHK1) 24 h after treatment with DMSO, NRH (100 μM), PARGi (10 μM) or NRH+PARGi. β-Actin was used as the loading control. (G) Immunoblot analysis of PAR and γ-H2AX in whole cell lysates prepared from GSC-83 cells treated with NRH (100 μM) + PARGi (10 μM) for 1 h under normal growth conditions, arrested by thymidine or released from thymidine arrest for 0.5, 1, 2, 4 or 6 h. β-Actin was used as the loading control.