Fig. 6.
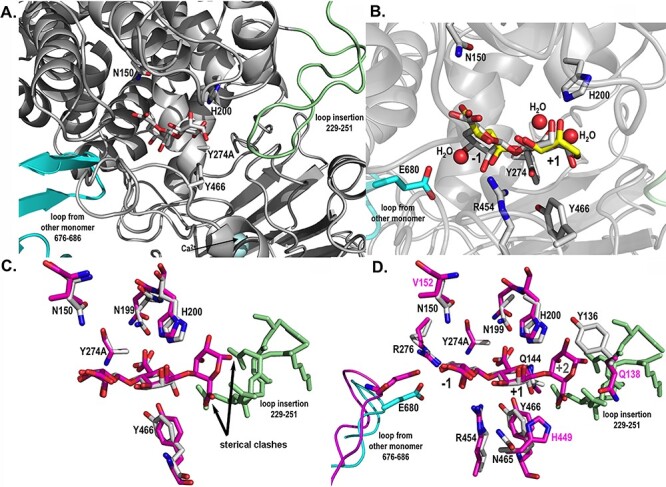
Catalytic active site structures of AlyA3 with bound molecules. (A) Close up view into the active site of AlyA3_Y274A, showing the relative position of a bound Δ-M substrate molecule with respect to the two loops blocking the entrance to the groove. The β-hairpin loop coming from the neighboring monomer B is colored in cyan, and the loop insertion, characteristic of AlyA3, is colored in light green. (B) Close up view into the active site of native AlyA3, showing the position of solvent molecules, malic acid and glycerol (carbons in yellow) that together with several water molecules (red spheres) occupy positions of the hydroxyl and acidic groups of the bound DP2 (Δ-M) substrate molecule (carbons in white) positioned here by superimposition with the AlyA3_Y274A complex with DP2 (panel A). Conserved and catalytic active residues are labeled. (C) Close up view into the active site of AlyA3_Y274A with Δ-M (carbons in white) superimposed with the DP3 molecule Δ-M-G bound in Alg17c (carbons in magenta). The figure highlights the sterical clashes, pointed out by the black arrows, of any additional guluronate unit at the potential +2 binding site with the inserted loop of AlyA3 (colored in light green). The atoms of the +2 guluronate unit would come to lie at 0.1 and 1.3 Å from atoms of the amino acids within the loop insertion. (D) Close up view into the active site of AlyA3_Y274A with Δ-M (carbons in white) superimposed with the DP3 molecule bound in Alg17c (carbons in magenta). The figure additionally displays the residues surrounding the substrate molecule and that differ between AlyA3 and Alg17c. Residue numbers in black label the residues of AlyA3, and labels in magenta highlight the residues that differ between the two enzymes, in close proximity of the catalytic site. This figure is available in black and white in print and in color at Glycobiology online.
